
DNA mediated self-assembly of multicellular microtissues
Introduction
Microphysiological systems (MPS) recently have emerged as a promising alternative for recapitulating the structure and function of native tissues in vitro (1). The emergence of MPS has sparked a rapidly growing interest within the tissue engineering and pharmaceutical industries due to its accelerating drug development and toxicity screening (2). However, engineering a tissue at the microscale level is challenged by differentiating stem cells self-assemble into the desired microphysiological structure. Lipid membranes define functional interfacial barriers for structural organization and compartmentalization within cells and supply a basic platform for tightly regulating various cellular events, such as energy conversion, intracellular transport, signal transduction, and cell communication (3,4). Consequently, DNA mediated self-assembly encouraging cell to communicate with one another may achieve a microphysiological system. In addition, mimicry of biology’s compartmentalization also holds great potential for technology development in nanoreactors, nanomedicine, and synthetic biology (5).
The past decades have witnessed the enormous development of DNA nanotechnology since the pioneering work of Nadrian Seeman on DNA nanostructures (6,7). By taking advantage of the unique, sequence-specific self-assembly properties of DNA, researchers can construct almost arbitrarily shaped nanoscale molecular structures (8-11). DNA nanostructures as rigid molecular scaffolds have also been exploited to precisely position molecules or other nanoscale objects after chemical functionalization (12,13). Moreover, DNA nanostructures can be employed to imitate protein structures, including cytoskeletal filaments (14), molecular cages (15) and enzyme scaffolds (16,17). However, some important features, especially large scale spatial organization and compartmentalization, will be missing when substituting proteins by DNA assembles.
Lipids, an essential class of biomolecules involving in biological compartmentalization, possess several intrinsic advantages. Such as, superstructures formed via self-assembly are intrinsically different from that formed by proteins or DNA; the lipid-lipid interactions are capable to form larger and extended molecular assemblies due to hydrophobic interactions (18). Notably, as aforementioned, membranes separate functionally distinct regions, while membrane-embedded molecules act as intermediary for intercompartmental communication and transport. Therefore, organization of functional molecules within membranes with a reduction in dimensionality would favor more effective interactions than in three dimensions.
On the basis of the significant progress in DNA nanotechnology, integrating DNA with lipid to construct DNA-lipid hybrid system becomes accessible in spite of their different chemical nature. As a result, various DNA-lipid conjugates have been developed and applied for realization of DNA-mediated self-assembly of multicellular MPS to study physicochemical and physiological features of membrane assemblies (19). Moreover, there artificial DNA-lipid hybrid systems with biocompatibility hold great promise as a powerful tool for versatile applications in drug delivery, synthetic biology, and chemical process control. Therefore, we give an overview of recent research progress and future developments of DNA-lipid hybrid systems in this review. In the following, we briefly introduce biophysical and biochemical characteristics of membranes. We then discuss membrane interactions with DNA, and assess properties of DNA-functionalized membrane and factors for membrane-anchored DNA. Finally, we summarize the applications of DNA-lipid hybrid systems, and give the outlook and challenges for future developments.
Physicochemical properties of cellular membranes
Lipids and their assemblies
As small organic molecules, lipids are soluble in nonpolar solvents but usually insoluble in water. Generally, there are three main categories of membrane lipids in nature—phospholipids, sphingolipids, and sterols (20). Phospholipids are composed of two hydrophobic acyl chains and one hydrophilic phosphate headgroup, which constitute the bulk of the membranes’ lipid matrix. Note that the structure of the phospholipid headgroup may be zwitterionic or anionic. In addition, composition of both acyl chain and headgroup can influence the physical properties of the membrane (21). Different from phospholipids, sphingolipids consist of trans-unsaturated or saturated acyl chains linked to a serine backbone, whose acyl-chain composition triggers the formation of a tall and narrow cylinder to increase its packing density in the membrane. As a result, a sphingomyelin bilayer exists in a solid gel phase with tightly packed, immobile acyl chains at physiological temperatures (22,23). They are also fluidized by sterols, which are major apolar membrane lipids with cholesterol predominating in mammals (24). Sterols interfere with acyl-chain packing and thus inhibit the transition of the membrane to the solid gel state. This reduces the flexibility of unsaturated acyl chains to rigidify fluid membranes, as a result increasing membrane thickness and impermeability to solutes (25-27). Hence, sterols assure that cells can minimize unregulated solute movement across membranes, while simultaneously keep them fluid.
Due to their amphipathic property, lipids can self-associate to form micelles or bilayer structures depending on their size and shape. The lipid structure organization in water derives from a compromise among steric hindrance, ionic repulsive forces, and hydrophobic association. Small and large unilamellar vesicles (SUVs and LUVs) with typical sized of 25–100 nm and ~1 µm form after hydration of dried lipid films, and applying electrical fields induces the formation of cell-sized giant unilamellar vesicles (GUVs). The curvature of lipid membranes is essential for the organization and compartmentalization of cells and cell organelles (28), and the changes in membrane curvature and topology reflect biological processes including budding and fusion of vesicles, or cell division (29). While a spontaneous curvature can be induced by an asymmetry in lipid distribution and the presence of membrane proteins, substantial forces supplied by specialized membrane-sculpting proteins could lead to large membrane deformations (30).
Lipid phases in membrane
Since membrane phase behaviors are correlated with the lipid-lipid interactions, the lipids of biological membranes display multiple possible phase states (31). It has been found that membranes are in a solid-ordered gel (Lβ) phase at low temperatures; whereas in a liquid-disordered phase (Lα) at high temperatures. The adopted phase relies on lipid structure: long, saturated hydrocarbon chains tend to adopt solid-like phases; while unsaturated hydrocarbon chains tend to adopt liquid phases. Several phases may also co-exist in a mixture of lipids. For instance, a liquid-ordered (Lo) phase distinct from Lβ may form when mixing cholesterol with phospholipids but cannot form without cholesterol (32). Consequently, the complex phase behavior is often used to mimic the external leaflet of the plasma membrane, and three-lipid mixtures containing cholesterol, sphingomyelin, and phosphatidylcholine lipids have been extensively studied (33-35). Taken together, the phase properties of membranes certainly play important roles in biological membrane function. And it should be pointed out that most membrane-associated proteins prefer liquid over solid phases. In addition, a majority of membrane proteins partition into the disordered phase because of coexistent Lo and Lα states.
Methods for DNA immobilized cellular membranes
Interactions between DNA and membranes
Electrostatic interactions
When liposomes are composed of cationic lipids [e.g., 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) or 1,2-di-O-octadecenyl-3-trimethylammonium propane (DOTMA) Figure 1A], a spontaneous electrostatic interaction between the positively charged lipids and the negatively charged DNA occurs (36). Their strong interaction causes an efficient condensation of DNA, thus forming spherical “lipoplexes” (37). Compared with electrostatic screening effects, the interaction between DNA and lipids is the strongest when the salt concentration is low (38). In order to surmount fragility of the artificial systems, Kurokawa et al. created a DNA shell underneath the membrane via exploiting cationic lipids to attract negatively charged DNA. They proved that the constructed DNA gel shell can stabilize the lipid membrane skin to the cytoskeleton in live cells (14).
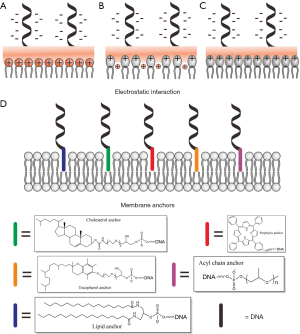
Zwitterionic lipids, including phosphatidylcholine and phosphatidylethanolamine, are incapable of condensing DNA into stable aggregates, while zwitterionic lipids are capable of binding strongly to DNA upon the addition of divalent metal cations such as Ca2+, Mg2+, or Mn2+ (39-41). The zwitterionic lipids possess a large headgroup dipole that is derived from the spatial separation of the negatively charged phosphate group from a positive charged moiety. A possible mechanism for the interaction with DNA is that binding of the divalent cations to the phosphate groups endows zwitterionic lipids with a net positive charge (Figure 1B) (39,40). Mengistu et al. proposed a continuum Poisson-Boltzmann formalism to explain the interaction of a zwitterionic lipid-DNA complex in the presence of divalent cations (42). However, the mechanism of the interaction has not been fully understood. Moreover, some studies have also revealed that DNA weakly binding to neutral lipids in the absence of the divalent cation (Figure 1C) may be mediated by ion-dipole interactions (43-45).
Electrostatic binding often depends on lipid composition and its phase behavior. Previous studies have verified that dsDNA preferred to bind to the Lo phase (46), which is attributed to the closer packing and thus higher surface charge density of the lipids.
Membrane anchors
Membrane anchors have been studied as the main approaches to binding DNA to membrane, which include lipid anchors, cholesterol anchors, porphyrin anchors, tocopherol anchors, and alkyl chains (Table 1).
(I) Lipid anchors
The highly negatively charged DNA-based nanostructures such as DNA nanopores, which are formed either by scaffold and staple strands (50) or short oligonucleotides (53,59), can also insert into hydrophobic bilayer membranes using DNA conjugates with lipids or other hydrophobic molecules (Figure 1D). Thus, lipid anchoring has been reported for bilayer-floating DNA nanostructure (51) and membrane-puncturing DNA nanopore (52). Given that DNA-lipid conjugates with relatively lower toxicity can induce vesicle fusion (47) or specially cross-link vesicles or cells (48), they have also been exploited for gen transfection (49). There are two approaches to synthesize lipid-DNA conjugates: a lipid tail group-like glycerol derivative is attached to any DNA sequence via phosphoramidite coupling during DNA synthesis; lipids are conjugated to amino- or thiol-modified DNA oligonucleotides after DNA synthesis. Note that the conjugation step can be directly carried out on a lipid membrane with reactive lipids (60).
(II) Cholesterol anchors
Despite their comparatively weak association with membranes, cholesterol may be one of the most studied lipids for DNA conjugation (50,51,61). Some studies reported that single cholesterol anchors could be sufficient to modify liposomes with 38-base DNA-aptamers for cell-specific targeting (62) and membrane-assisted assembly of DNA structures (63). On the other hand, cholesterol anchors might cause nearly irreversible binding to membranes (64-66), despite that multiple cholesterol modifications result in enhanced hydrophobic association with membranes (64). Walter group reported that cholesterol-labeled DNA origami barges are capable of reversible association with and lateral diffusion on lipid bilayers and the assembled DNA barges embedding in lipid bilayers present Brownian diffusion in a manner relying on cholesterol labeling and bilayer composition (67).
The binding properties of cholesterol depend on the nature of the linker. Individual cholesteryl-anchored ssDNA inserts into lipid vesicles or supported lipid bilayers, and their conformation becomes more rigid with increasing concentration, which influenced hybridization kinetics with complementary oligonucleotides (68). Note that cholesterol linked to tetraethylene glycol (chol-TEG), compared with cholesterol alone, neither alters the bilayer structure and dynamics of membranes nor induces condensation of the membrane lipids (68). Beales et al. (69) investigated the partitioning of different cholesterol-modified ssDNA molecules (chol-DNAs) between the domains of phase-separated lipid vesicles. They found that chol-DNAs with single cholesterol anchor partitioned roughly equally between coexisting domains; while chol-DNAs with two cholesterol anchors preferred the Lo over the Lα phase. On the other hand, chol-TEG-DNA conjugates partitioned into the Lα phase only.
(III) Porphyrin anchors
Owing to its similar size, shape and hydrophobicity to cholesterol, the porphyrin and its derivative with strong anchoring force show potential physical interaction in the lipid membrane (53). Moreover, porphyrin also has a high van der Waals surface area, which can be further increased with additional aromatic substituents. In recent years, several groups have synthesized porphyrin-DNA conjugates and thus constructed supramolecular system anchored to liposomes or membranes for further study (54). On the basis of these, Börjesson et al. (54) found that the porphyrin, in contrast to the inert cholesterol anchor commonly used, is well suited as a photophysical and redox-active lipid anchor. Meanwhile, porphyrins are often incorporated into DNA strands using standard solid phase phosphoramidite oligonucleotide synthesis techniques to synthesize porphyrin-DNA conjugates (70-72). It is worth to point out that porphyrins acting as powerful visualization tags provide a handle to verify membrane anchoring when inserting porphyrins into lipid bilayers results in characteristic shifts in their fluorescence spectrum (54,73).
(IV) Tocopherol anchors
Recently, α-tocopherol also has been reported to be attached to DNA (55). For instance, α-tocopherol-modified oligonucleotides can spontaneously inset into pre-formed lipid vesicles within seconds and prefer to incorporate into Lα phase. This process does not cause significant perturbation of the lipid bilayers, suggesting such DNA-α-tocopherol conjugates are suitable for functionalization of biological membranes (55). Furthermore, Takeshi Tokunaga et al. demonstrated that DNA conjugated to α-tocopherol at the terminal phosphate moiety could spontaneously incorporate into cell membranes (74). In addition, Andreas Herrmann group found that α-tocopherol units as lipophilic anchors could stabilize the double-helix formation in distinct membrane domains (75).
(V) Alkyl chains
Apart from above anchors, alkyl chains have been considered as an effective anchor tool to bind DNA to membranes (76). Therefore, conjugates of oligonucleotides with poly (propylene oxide) (77) or fatty acids (56) have been reported to incorporate into lipid membranes, wherein DNA backbone become hydrophobic (57). Sleiman group synthesized dendritic alkyl-DNA conjugates hybridized to the edges of a DNA cube allowed for encapsulation and release of hydrophobic small molecule cargo (58).
Membrane-bound proteins
DNA may also interact with membranes through membrane-associated proteins, which are often overexpressed on the surface of a certain cell type or some diseased cell. For example, human protein tyrosine kinase 7 is overexpressed on CEM (human T-cell acute lymphocytic leukemia) cell membrane (78). Consequently, this characteristic can be exploited to target specific biological membranes.
In summary, there are several approaches which can achieve binding to membrane proteins: DNA-antibody conjugates (79), aptamer-target interactions (80) and covalent coupling (81). Nevertheless, these approaches distinct from other interactions mentioned above, does not need to involve a direct interaction with the lipid membrane itself (18).
Functional membrane-anchored DNA
Properties of membrane-anchored DNA
Similar to free DNA oligonucleotides, membrane-anchored DNA oligonucleotides are capable to bind to complementary DNA from aqueous environment. Differently, its rate constant of oligonucleotide hybridization, compared to that in solution, is faster at low membrane grafting, but is slower at higher density (82). This principle can thus be used to control the processes of higher complexity when complementary interactions of two populations of liposomes with various sizes are mediated by cholesteryl-oligonucleotide (19). Besides, DNA hybridization is an effective approach to reveal essential physical mechanisms underlying vesicle-vesicle interactions as well. In general, adhesion plaques between pairs of liposomes saturate at about 20 DNA molecules per vesicle, above which excess DNA molecules bind preferentially to rest liposome, resulting in the formation of liposomal precipitates. Furthermore, more complementary membrane-anchored DNA could achieve multicompartment cluster composed of different liposomes (83). Accordingly, multilayer aggregates with each layer consisting of a unique population of membrane-embedded compartments can organize and regulate biochemical reactions at the nanoscale (84), wherein the reversible crosslinking may be attributed to hydrophobic anchors-modified DNA strands at both ends (85). On the other hand, when ssDNA inserts both lipophilic anchors into the same vesicles, the rigidity of the double helix increases and membrane anchor is released into solution. In summary, such reversible assembly of liposomes provides a precise switch between assembled and disassembled states of multi-compartmentalized membranous systems (86).
Membrane-anchored DNA oligonucleotide hybridization can also be employed to investigate the phenomena associated with ligand-receptor docking and membrane-surface tethering. Previous study revealed that modeling lateral domain organization of membrane surface receptors and cell-to-cell junctions is implicated by lipophilic DNA-anchored membranes on top of lipid bilayers; while the lateral mobile lipid-DNA conjugates tend to segregate by their height (8–24 nm) and the upper lipid bilayer can accommodate the height difference via deformation (87,88). On the basis of these findings, Chan et al. exploited DNA-tethered vesicles attached to supported lipid bilayers for displaying membrane components (89). Importantly, this novel strategy provides a unique possibility to conduct many studies, for instance, single particle tracking analysis of diffusion before and after docking. Note that DNA-anchored membranes lower the dimensionality of a studied system, which facilitates monitoring membrane-associated events using sensitive surface techniques [e.g., total internal reflection fluorescence microscopy (90), quartz crystal microbalance (91), surface plasmon resonance (92), and fluorescence recovery after photobleaching (93)]. Hence, supported lipid bilayer-mediated assembly of liposomes serves as a useful tool to create and mimic complex multicompartment assemblies.
Factors for membrane-anchored DNA
There are several factors to consider regarding DNA binding to lipid membranes. One key factor is the lipid composition. Beales et al. investigated the thermal stability of DNA duplex formation using membrane-anchored DNA as model adhesion receptors between lipid vesicles. They found that the melting temperature of DNA sequence with short bases is strongly dependent on the lipid charge of the vesicles membranes (94). Apart from lipid charge, the phase properties are also important factors that affect membranes binding to DNA because of their contribution to physiological function (95,96). As mentioned above, membranes present multiple possible phase states, including Lα, Lβ, and Lo phase. Here, membrane-anchored DNA preferentially partitions between coexisting phases, giving rise to the appearance of domains with distinct functionalities. A similar behavior was also reported in previous work (55,75), in which GUVs were localized in the Lα domains. Moreover, the DNA sequences are another important factor. To study the effect of membrane-membrane spacing on fusion, Boxer group constructed a series of conjugates with 2–24 non-complementary bases at the membrane-proximal ends of two complementary sequence. They found that effects of linker sequence on vesicle fusion could be mediated by lipid-anchored DNA oligonucleotides (97,98). The rates and extents of lipid and content mixing are progressively reduced with increasing linker lengths. In addition, Gartner demonstrated that the kinetic parameters of the assembly of 3-dimensional microtissues with defined cellular connectivity depend on DNA sequence complexity, density, and cell concentration (99).
DNA-mediated self-assembly of artificial multicellular systems
Biomimetic trans-membrane channels
With the development of DNA nanotechnology, more sophisticated DNA origami-based synthetic systems, such as artificial lipid bilayer-embedded channels, have been constructed by insertion of DNA nanostructures into a lipid bilayer, which mimics the behavior of membrane proteins to control the transport of molecules and ions. Through the interactions between DNA and membranes described above, the DNA nanostructures can overcome the large free energy penalty and are inserted into lipid, thus forming a hole in the membranes. In addition, such synthetic membrane channels, compared to the connexins within gap junctions between cells (100), bacterial drug efflux transporters (101), and nuclear pore complex (102), offer a better route to engineer the diameter of channels using DNA nanotechnology (5). Furthermore, the innovative studies have also demonstrated that DNA origami-based transmembrane channels have electrophysiological properties similar to membrane proteins (50,59,103).
Langecker et al. designed channel-like engineered nanostructures composed of 54 parallel DNA helices, of which six rounding up the center penetrates through the membrane (Figure 2A) (50). As shown in Figure 2B, TEM image verifies that these mimetic structures are inserted into membranes with the desired orientation. This bacterial toxin α-hemolysin-inspired channel is anchored to the lipid bilayer by 26 cholesteryl anchors. Consequently, the channels are endowed with a strong hydrophobic association with the membrane, which compels the inner channel to penetrate through the membrane. However, the penetrating column does not contain any hydrophobic modifications causing lipid bilayer rearranged, as a result a hydrophilic pore around the DNA channel forms. These features make the mimetic channels yield gating properties similar to natural ion channels. This innovated work has taken the first step toward harnessing ion flux for driving sophisticated nanodevices.
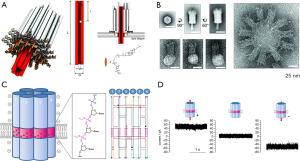
Another example of mimetic channel was based on hydrophobic modified structures on the outer surface of the transmembrane part matching the hydrophobic thickness of the membrane. Burns and co-workers developed a DNA-based nanopore with six interconnected DNA duplexes of about 15 nm long, which was designed to form a central pore of around 2 nm (Figure 2C) (59). In this case, ethyl modified phosphorothioate group instead of phosphate group was used for targeted chemical modification of DNA backbones, which exactly matches the thickness of the lipid bilayer. This artful design enabled a stable DNA channel insertion into the membrane and a tight seal with surrounding lipid environment (53). When a potential of +100 mV (0 or −100 mV) was applied to DNA-nanopore, a constant flow of ionic current was generated (Figure 2D), validating that the DNA-nanopore can work as trans-membrane channels. Such membrane-spanning DNA channels hold great promising to aid the design of new molecular devices for controlled transmembrane transport (104).
DNA structures for synthetic biology
It is well acknowledged that lipid bilayers function as a dynamic boundary around cells and subcellular organelle units, which inspires scientists to prepare artificial vesicles or liposomes as model systems to investigate cell biology and drug carriers interfering with cell behavior (105,106). However, compared to proteins or DNA, membranes lack a defined structure, thus resulting in fragile lipid membranes. In biology, cytoskeleton which is made up of interconnected protein rods linked to the membrane is utilized to shape and stabilize membranes. Nevertheless, the cytoskeleton is difficult to engineer due to its complex architecture, and to design predictable changes in structure. It is envisioned that DNA nanotechnology provides an ideal tool for synthetic biology. As a result of sequence-specific and directional hybridization of DNA duplexes, it is easy to fold and assemble DNA strands into structures with well-defined size and shape. Moreover, in contrast with proteins, the complexity of DNA sequence is greatly reduced with easy design procedures. Taking these features into consideration, it is possible to construct nanoscale objects with a designed shape using DNA nanotechnology.
Inspired by the protein machineries sculpting and scaffolding membrane structures in cells (107), Yang and co-workers designed four different DNA-origami rings and used them as templates to define the liposome size (Figure 3A) (108). First, they exploited structural DNA nanotechnology (110) to produce self-assembled nanostructures with addressable surface, programmable geometry, and outstanding structural stability as nanotemplates for vesicles, via the rational design of DNA-strand hybridization. Following, the interior surface of the prepared DNA origami nanostructures was decorated with lipid molecules, which was used to guide the formation of liposomes. Finally, a series of highly monodispersed liposomes with sizes of 29, 46, 60 and 94 nm were yielded (Figure 3B). In comparison with other artificial templates surrounded by membranes (i.e., an endoskeleton) (111), the DNA nanostructure developed by Lin functions as the exoskeleton and generates vesicles with more versatile functions. Meanwhile, this system provides a good example for nucleated lipid assembly into a liposome within a DNA nanostructure template.
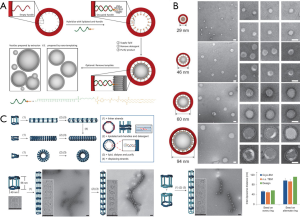
Recently, Lin group have applied various advanced fabrication techniques, including caDNAno (112), curved multi-helix bundle (113), shape complementarity (114), and flexible connection between stiff components (115), for designing DNA origami nanocages as templates to guide liposome formation (Figure 3C) (109). Each cylinder-like nanocage consists of two DNA rings separated by four pillars with connector sites to assemble multiple cages. After heating and cooling a mixture of DNA strands with specific sequences, the nanocages were assembled into linear nanoscaffolds. Thereafter, phospholipid anchors were tethered to the cages’ DNA rings to position liposomes inside the cages, followed by adding excess free lipids to induce the formation of liposomes via self-assembly. Notably, this modular design with tunable parameters, including the ring size, the pillar length and rigidity, the number and position of the handles and teeth, allows to construct different frames. This modularly designed DNA nanocages thus produced high-order nanostructures that can serve as templates for different bilayer shapes (116).
Drug delivery/nanomedicine
Another promising application of DNA nanostructures in the field of lipid membranes is to develop hybrid delivery systems for biomedical purposes. Extensive studies on cellular uptake and intracellular stability of DNA-based nanostructures have been carried out (117). A phenomenological work by Church group developed a logic-gated nanorobot for targeted transport of molecular payloads (Figure 4A) (79). The system is composed of a barrel with two domains, which are attached by single-strand scaffold hinges on the one side and fastened by molecular locks on the other side. Inspired by aptamer beacons and structure-switching aptamers, they designed DNA aptamer-based locks responsive to antigen keys. Consequently, the aptamers bind preferentially to their targets in the presence of antigen keys, and thus unlock the barrel, resulting in the release of molecular payload. Moreover, they took advantage of TME imaging to demonstrate DNA nanodevice in closed and open states, with and without molecular payload (Figure 4B). The work provides a prototype for new designs with selective delivery of biologically active payloads for cell-targeting tasks.
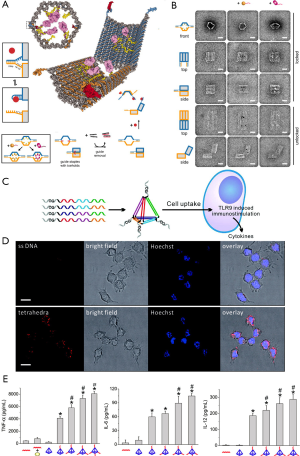
It has been demonstrated that the cytosine-phosphate-guanine (CpG) oligonucleotides can be recognized by Toll-like receptor 9 and thus induce immune response (119). Therefore, CpG oligonucleotides are widely used as a potent activator in the immunotherapy. However, the delivery of CpG oligonucleotides remains challenging because of its easy degradation in physiological conditions and difficulty to enter cells and reach the target sites (120). To address these obstacles, Li and co-workers constructed a DNA tetrahedron nanostructure as a multivalent nanocarrier for CpG. This DNA tetrahedron nanostructure is compact, mechanically stable, noncytotoxic, and resistant to nuclease degradation (Figure 4C) (118). Moreover, these CpG-tetrahedron nanostructures, compared with ssDNA, showed higher efficiency to enter macrophage-like RAW264.7 cells (Figure 4D) and stimulated higher secretion levels of certain cytokines (Figure 4E). The work provides unprecedented opportunities to design drug delivery nanocarriers capable to tune the drug dosage.
Membrane fusion
Membrane fusion, one of the most fundamental processes, is essential for eukaryotic cell function. It is coordinated by membrane-anchored fusion proteins, for instance like N-ethyl-maleimide-sensitive-factor attachment protein (SNARE) (121), wherein a specific complex formed by the cytosolic domains of these proteins pulls the membranes into close. However, there is a great controversy in the mechanism underlying fusion (122-124). To mimic SNARE fusion protein function (125), Gudrun Stengel and co-worker built a fusion machinery composed of cholesterol-modified DNA zippers in a reductionist approach to initiate vesicle fusion (Figure 5A) (47). Here, the cholesterol-anchored DNA structure aims to minimizing the distance between two DNA-bridged bilayers. On the other hand, DNA hybridization in a zippering action stars at the membranes distal ends and proceeds to the membrane proximal ends, pulling the membranes into close apposition. In addition, they explored the effect of anchoring strategy, DNA length and DNA surface coverage on DNA-mediated fusion. This work mimics SNARE protein function and broadens the application of recognition guided membrane fusion.
Fluorescence assays are generally employed to study the membrane fusion, such as lipid mixing, inner monolayer lipid mixing, and content mixing (97,127,128). Previous study revealed that lipid mixing initiated by the DNA-zippering mechanism can be up to ~80% (129) and its efficiency are affected by both lipid compositions and DNA properties. For example, Gudrun Stengel et al. found that the rate and extend of lipid mixing between liposomes can be amplified by inverted-cone shaped lipids (127). As a result, these lipids induced the increasing of the stored curvature elastic stress within membranes and lowered the free energy barrier to the topological changes (130).
Mimicking membrane-related biological events
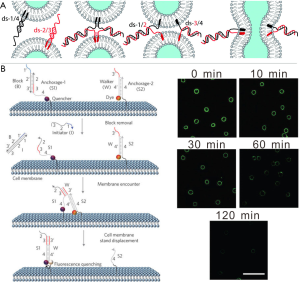
The membrane mediates various fundamental biological processes, such as molecular transport, energy conversion, signal transduction, and intercellular communication (131). Note that disease progression is induced upon disruption of such membrane-bound interactions or encounters (132). It is therefore of great importance to investigate insight of interaction patterns on live cell membranes. However, it remains a technical challenge to monitor the interactions at the cell surface, because of the short lifetimes (µs–ms) (133) of transient signaling encounter events. Recently, You group developed a novel DNA probe to address this challenge by transducing transient membrane encounter events into readable cumulative fluorescence signals (Figure 5B) (126).
You and co-workers constructed a well-regulated dynamic DNA system by employing the live cell membranes and membrane compounds as the track (T) and anchor sites (S), respectively. Inspired by tiny motor proteins powering locomotion (134), the prepared DNA probes steps onto S and moves along a T (135). Here, toehold-mediated DNA strand displacement (136) was employed to make DNA probe (W) translocation from one anchor (S2) to another (S1), in which W hybridizes to S1 resulting in the displacement of S2 from the S2/W conjugate with the formation of S1/W conjugates. It is worth to mention that the strand exchange rate exhibited a linear correlation with the rate of anchor sit encounter on the membrane. Therefore, the encounter dynamics of two anchor sited can be calculated through monitoring the time-dependent variation of DNA probe translocation. As shown in Figure 5B, the changes in fluorescence intensity was monitored by fluorescence microscopy, thus confirming the membrane encounter dynamics. This strategy combined with compositional profiling of the membrane provides a powerful tool to aid understanding biological membranes and their role in health and disease (137).
Outlook and challenges
The remarkable self-assembly property of DNA broadens its biological applications. DNA nanotechnology as a rapidly evolving tool shows unique advantages to engineer biomolecules and biologically relevant processes at the nanoscale. Accordingly, interacting DNA or DNA nanostructures with lipids to construct DNA-lipid hybrid systems opens up a new approach to study and analyze many biologically important issues, including molecular transport and cell communications. Many biological events thus have been successfully reproduced with the aid of DNA-lipid hybrid systems owing to their spatial organization of ligands and receptors, membrane curvature, and phase segregation in a controllable manner, whereas it is impossible to achieve this using conventional methods. In order to gain deeper insight into interaction of DNA with lipids and broad the applications of DNA-lipid hybrid systems, nevertheless, several major challenges must be addressed. First, developing a general framework allows to program the interconnections and superstructures composed of arbitrary numbers of liposome populations, which goes beyond conventional binary systems. To achieve this goal, theoretical developments to assemble complex and multicomponent structures from colloidal particles may be a promising starting point (138). Another challenge is how to efficiently control chemicals transport via responsive gating mechanisms, including compartments with chemical specificity and the possibility of controlling transport. It would be a significant step to control trans-membrane transport by virtue of current techniques, because non-specific nanopores and complete content mixing or release are commonly studied in synthetic systems. With regards to their potential drug delivery application, more attention should be focused on understanding the interactions of lipid-DN A hybrid systems with cells and even whole organisms in order to assess their viability. Considering the versatility of DNA-lipid hybrid systems, it is envisioned that future studies may not be restricted to lipid-based confining layers and will exploit DNA-mediated assembly for constructing more robust polymersomes (139), hybrid vesicles (140) and lipo-polymersomes (141).
Acknowledgments
Funding: This work was supported by the National Science Foundation of China (grant numbers 21722502, 21505045, 21705048), the Shanghai Pujiang Talent Project (16PJ1402700), China Postdoctoral Science Foundation (2015M581565, 2017T100283).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/mps.2017.12.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ewart L, Dehne EM, Fabre K, et al. Application of microphysiological systems to enhance safety assessment in drug discovery. Annu Rev Pharmacol Toxicol 2017;58:65-82. [PubMed]
- Zhang YS. Foreword—Microphysiological Systems. Microphysiol Syst 2017;1:1.
- Di Paolo G, De Camilli P. Phosphoinositides in cell fegulation and mmembrane dynamics. Nature 2006;443:651-7. [Crossref] [PubMed]
- Gumbiner BM. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell 1996;84:345-57. [Crossref] [PubMed]
- Beales PA, Vanderlick TK. Application of nucleic acid–lipid conjugates for the programmable organisation of liposomal modules. Adv Colloid Interface Sci 2014;207:290-305. [Crossref] [PubMed]
- Seeman NC. De novo design of sequences for nucleic acid structural engineering. J Biomol Struct Dyn 1990;8:573-81. [Crossref] [PubMed]
- Chen JH, Seeman NC. Synthesis from DNA of a molecule with the connectivity of a cube. Nature 1991;350:631-3. [Crossref] [PubMed]
- Pinheiro AV, Han D, Shih WM, et al. Challenges and opportunities for structural DNA nanotechnology. Nat Nanotechnol 2011;6:763-72. [Crossref] [PubMed]
- Seeman NC. DNA in a material world. Nature 2003;421:427-31. [Crossref] [PubMed]
- Pei H, Zuo XL, Pan D, et al. Scaffolded biosensors with designed DNA nanostructures. NPG Asia Mater 2013;5:e51 [Crossref]
- Abi A, Lin MH, Pei H, et al. Electrochemical switching with 3D DNA tetrahedral nanostructures self-assembled at gold electrodes. ACS Appl Mater Interfaces 2014;6:8928-31. [Crossref] [PubMed]
- Schreiber R, Do J, Roller E-M, et al. Hierarchical assembly of metal nanoparticles, quantum dots and organic dyes using DNA origami scaffolds. Nat Nanotechnol 2014;9:74-8. [Crossref] [PubMed]
- Chen Q, Liu HJ, Lee W, et al. Self-assembled DNA tetrahedral optofluidic lasers with precise and tunable gain control. Lab Chip 2013;13:3351-4. [Crossref] [PubMed]
- Kurokawa C, Fujiwara K, Morita M, et al. DNA cytoskeleton for stabilizing artificial cells. Proc Natl Acad Sci U S A 2017;114:7228-33. [Crossref] [PubMed]
- Zhang C, Su M, He Y, et al. Conformational flexibility facilitates self-assembly of complex DNA nanostructures. Proc Natl Acad Sci U S A 2008;105:10665-9. [Crossref] [PubMed]
- Zhao Z, Fu J, Dhakal S, et al. Nanocaged enzymes with enhanced catalytic activity and increased stability against protease digestion. Nat Commun 2016;7:10619. [Crossref] [PubMed]
- Shen J, Xu L, Wang C, et al. Dynamic and quantitative control of the DNA-mediated growth of gold plasmonic nanostructures. Angew Chem Int Ed Engl 2014;53:8338-42. [Crossref] [PubMed]
- Langecker M, Arnaut V, List J, et al. DNA nanostructures interacting with lipid bilayer membranes. Acc Chem Res 2014;47:1807-15. [Crossref] [PubMed]
- Beales PA, Vanderlick TK. Specific binding of different vesicle populations by the hybridization of membrane-anchored DNA. J Phys Chem A 2007;111:12372-80. [Crossref] [PubMed]
- Holthuis JC, Menon AK. Lipid landscapes and pipelines in membrane homeostasis. Nature 2014;510:48-57. [Crossref] [PubMed]
- de Kroon AI, Rijken PJ, De Smet CH. Checks and balances in membrane phospholipid class and acyl chain homeostasis, the yeast perspective. Prog Lipid Res 2013;52:374-94. [Crossref] [PubMed]
- Slotte JP. Biological functions of sphingomyelins. Prog Lipid Res 2013;52:424-37. [Crossref] [PubMed]
- Qu X, Yang F, Chen H, et al. Bubble-mediated ultrasensitive multiplex detection of metal ions in three-dimensional DNA nanostructure-encoded microchannels. ACS Appl Mater Interfaces 2017;9:16026-34. [Crossref] [PubMed]
- Yeagle PL. The membranes of cells. London: Academic Press, 2016.
- Brown DA, London E. Structure and origin of ordered lipid domains in biological membranes. J Membr Biol 1998;164:103-14. [Crossref] [PubMed]
- Qu X, Zhang H, Chen H, et al. Convection-driven pull-down assays in nanoliter droplets using scaffolded aptamers. Anal Chem 2017;89:3468-73. [Crossref] [PubMed]
- Su S, Zuo XL, Pan D, et al. Design and applications of gold nanoparticle conjugates by exploiting biomolecule-gold nanoparticle interactions. Nanoscale 2013;5:2589-99. [Crossref] [PubMed]
- Phillips R, Ursell T, Wiggins P, et al. Emerging roles for lipids in shaping membrane-protein function. Nature 2009;459:379-85. [Crossref] [PubMed]
- Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol 2006;7:9-19. [Crossref] [PubMed]
- McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol 2011;12:517-33. [Crossref] [PubMed]
- van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 2008;9:112-24. [Crossref] [PubMed]
- Marsh D. Cholesterol-induced fluid membrane domains: a compendium of lipid-raft ternary phase diagrams. Biochim Biophys Acta 2009;1788:2114-23.
- Veatch SL, Keller SL. Miscibility phase diagrams of giant vesicles containing sphingomyelin. Phys Rev Lett 2005;94:148101 [Crossref] [PubMed]
- Qu X, Wang S, Ge Z, et al. Programming cell adhesion for on-chip sequential boolean logic functions. J Am Chem Soc 2017;139:10176-9. [Crossref] [PubMed]
- Yang X, Li J, Pei H, et al. Pattern recognition analysis of proteins using DNA-decorated catalytic gold nanoparticles. Small 2013;9:2844-9. [Crossref] [PubMed]
- Pedroso de Lima MC, Simões S, Pires P, et al. Cationic lipid–DNA complexes in gene delivery: from biophysics to biological applications. Adv Drug Deliv Rev 2001;47:277-94. [Crossref] [PubMed]
- Felgner PL, Gadek TR, Holm M, et al. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc Natl Acad Sci U S A 1987;84:7413-7. [Crossref] [PubMed]
- Pozharski E, MacDonald RC. Thermodynamics of cationic lipid-DNA complex formation as studied by isothermal titration calorimetry. Biophys J 2002;83:556-65. [Crossref] [PubMed]
- McManus JJ, Rädler JO, Dawson KA. Does calcium turn a zwitterionic lipid cationic? J Phy Chem B 2003;107:9869-75. [Crossref]
- Gromelski S, Brezesinski G. DNA condensation and interaction with zwitterionic phospholipids mediated by divalent cations. Langmuir 2006;22:6293-301. [Crossref] [PubMed]
- Qi L, Xiao M, Wang F, et al. Poly-cytosine-mediated nanotags for SERS detection of Hg2+. Nanoscale 2017;9:14184-91. [Crossref] [PubMed]
- Mengistu DH, Bohinc K, May S. Binding of DNA to zwitterionic lipid layers mediated by divalent cations. J Phys Chem B 2009;113:12277-82. [Crossref] [PubMed]
- Ainalem ML, Kristen N, Edler KJ, et al. DNA binding to zwitterionic model membranes. Langmuir 2010;26:4965-76. [Crossref] [PubMed]
- Chen L, Chao J, Qu X, et al. Probing cellular molecules with PolyA-based engineered aptamer nanobeacon. ACS Appl Mater Interfaces 2017;9:8014-20. [Crossref] [PubMed]
- Yao G, Li J, Chao J, et al. Gold-nanoparticle-mediated jigsaw-puzzle-like assembly of supersized plasmonic DNA origami. Angew Chem Int Ed Engl 2015;54:2966-9. [Crossref] [PubMed]
- Kato A, Tsuji A, Yanagisawa M, et al. Phase separation on a phospholipid membrane inducing a characteristic localization of DNA accompanied by its structural transition. J Phy Chem Lett 2010;1:3391-5. [Crossref]
- Stengel G, Simonsson L, Campbell RA, et al. Determinants for membrane fusion induced by cholesterol-modified DNA zippers. J Phys Chem B 2008;112:8264-74. [Crossref] [PubMed]
- Selden NS, Todhunter ME, Jee NY, et al. Chemically programmed cell adhesion with membrane-anchored oligonucleotides. J Am Chem Soc 2012;134:765-8. [Crossref] [PubMed]
- Burns JR, Al‐Juffali N, Janes SM, et al. Membrane‐spanning DNA nanopores with cytotoxic effect. Angew Chem Int Ed Engl 2014;53:12466-70. [PubMed]
- Langecker M, Arnaut V, Martin TG, et al. Synthetic lipid membrane channels formed by designed DNA nanostructures. Science 2012;338:932-6. [Crossref] [PubMed]
- Czogalla A, Kauert DJ, Franquelim HG, et al. Amphipathic DNA origami nanoparticles to scaffold and deform lipid membrane vesicles. Angew Chem Int Ed Engl 2015;54:6501-5. [Crossref] [PubMed]
- Burns JR, Seifert A, Fertig N, et al. A biomimetic DNA-based channel for the ligand-controlled transport of charged molecular cargo across a biological membrane. Nat Nanotechnol 2016;11:152-6. [Crossref] [PubMed]
- Burns JR, Göpfrich K, Wood JW, et al. Lipid‐bilayer‐spanning DNA nanopores with a bifunctional porphyrin anchor. Angew Chem Int Ed Engl 2013;52:12069-72. [Crossref] [PubMed]
- Börjesson K, Wiberg J, El-Sagheer AH, et al. Functionalized nanostructures: redox-active porphyrin anchors for supramolecular DNA assemblies. ACS Nano 2010;4:5037-46. [Crossref] [PubMed]
- Bunge A, Kurz A, Windeck AK, et al. Lipophilic oligonucleotides spontaneously insert into lipid membranes, bind complementary DNA strands, and sequester into lipid-disordered domains. Langmuir 2007;23:4455-64. [Crossref] [PubMed]
- Borisenko GG, Zaitseva MA, Chuvilin AN, et al. DNA modification of live cell surface. Nucleic Acids Res 2009;37:e28 [Crossref] [PubMed]
- Gut IG, Beck S. A procedure for selective DNA alkylation and detection by mass spectrometry. Nucleic Acids Res 1995;23:1367-73. [Crossref] [PubMed]
- Edwardson TG, Carneiro KM, McLaughlin CK, et al. Site-specific positioning of dendritic alkyl chains on DNA cages enables their geometry-dependent self-assembly. Nat Chem 2013;5:868-75. [Crossref] [PubMed]
- Burns JR, Stulz E, Howorka S. Self-assembled DNA nanopores that span lipid bilayers. Nano Lett 2013;13:2351-6. [Crossref] [PubMed]
- Dave N, Liu J. Programmable assembly of DNA-functionalized liposomes by DNA. ACS Nano 2011;5:1304-12. [Crossref] [PubMed]
- Ge ZL, Pei H, Wang LH, et al. Electrochemical single nucleotide polymorphisms genotyping on surface immobilized three-dimensional branched DNA nanostructure. Sci China Chem 2011;54:1273. [Crossref]
- Cao Z, Tong R, Mishra A, et al. Reversible cell‐specific drug delivery with aptamer‐functionalized liposomes. Angew Chem Int Ed Engl 2009;48:6494-8. [Crossref] [PubMed]
- Bombelli FB, Betti F, Gambinossi F, et al. Closed nanoconstructs assembled by step-by-step ss-DNA coupling assisted by phospholipid membranes. Soft Matter 2009;5:1639-45. [Crossref]
- Pfeiffer I, Höök F. Bivalent cholesterol-based coupling of oligonucletides to lipid membrane assemblies. J Am Chem Soc 2004;126:10224-5. [Crossref] [PubMed]
- Qu X, Zhu D, Yao G, et al. An exonuclease III-powered, on-particle stochastic DNA walker. Angew Chem Int Ed Engl 2017;56:1855-8. [Crossref] [PubMed]
- Zhu D, Song P, Shen JW, et al. PolyA-mediated DNA assembly on gold nanoparticles for thermodynamically favorable and rapid hybridization analysis. Anal Chem 2016;88:4949-54. [Crossref] [PubMed]
- Johnson-Buck A, Jiang S, Yan H, et al. DNA–cholesterol barges as programmable membrane-exploring agents. ACS Nano 2014;8:5641-9. [Crossref] [PubMed]
- Bunge A, Loew M, Pescador P, et al. Lipid membranes carrying lipophilic cholesterol-based oligonucleotides–characterization and application on layer-by-layer coated particles. J Phys Chem B 2009;113:16425-34. [Crossref] [PubMed]
- Beales PA, Vanderlick TK. Partitioning of membrane-anchored DNA between coexisting lipid phases. J Phys Chem B 2009;113:13678-86. [Crossref] [PubMed]
- Balaz M, Holmes AE, Benedetti M, et al. Synthesis and circular dichroism of tetraarylporphyrin−oligonucleotide conjugates. J Am Chem Soc 2005;127:4172-3. [Crossref] [PubMed]
- Qu X, Li M, Zhang H, et al. Real-time continuous identification of greenhouse plant pathogens based on recyclable microfluidic bioassay system. ACS Appl Mater Interfaces 2017;9:31568-75. [Crossref] [PubMed]
- Pei H, Zuo XL, Zhu D, et al. Functional DNA nanostructures for theranostic applications. Acc Chem Res 2014;47:550-9. [Crossref] [PubMed]
- Qi L, Xiao M, Wang X, et al. DNA-encoded raman-active anisotropic nanoparticles for microRNA detection. Anal Chem 2017;89:9850-6. [Crossref] [PubMed]
- Tokunaga T, Namiki S, Yamada K, et al. Cell surface-anchored fluorescent aptamer sensor enables imaging of chemical transmitter dynamics. J Am Chem Soc 2012;134:9561-4. [Crossref] [PubMed]
- Kurz A, Bunge A, Windeck AK, et al. Lipid‐anchored oligonucleotides for stable double‐helix formation in distinct membrane domains. Angew Chem Int Ed Engl 2006;45:4440-4. [Crossref] [PubMed]
- Loew M, Springer R, Scolari S, et al. Lipid domain specific recruitment of lipophilic nucleic acids: a key for switchable functionalization of membranes. J Am Chem Soc 2010;132:16066-72. [Crossref] [PubMed]
- Rodríguez-Pulido A, Kondrachuk AI, Prusty DK, et al. Light-triggered sequence-specific cargo release from DNA block copolymer-lipid vesicles. Angew Chem Int Ed Engl 2013;52:1008-12. [Crossref] [PubMed]
- Zhu G, Zheng J, Song E, et al. Self-assembled, aptamer-tethered DNA nanotrains for targeted transport of molecular drugs in cancer theranostics. Proc Natl Acad Sci U S A 2013;110:7998-8003. [Crossref] [PubMed]
- Douglas SM, Bachelet I, Church GM. A logic-gated nanorobot for targeted transport of molecular payloads. Science 2012;335:831-4. [Crossref] [PubMed]
- Tang Z, Shangguan D, Wang K, et al. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal Chem 2007;79:4900-7. [Crossref] [PubMed]
- Hsiao SC, Shum BJ, Onoe H, et al. Direct cell surface modification with DNA for the capture of primary cells and the investigation of myotube formation on defined patterns. Langmuir 2009;25:6985-91. [Crossref] [PubMed]
- Banchelli M, Betti F, Berti D, et al. Phospholipid membranes decorated by cholesterol-based oligonucleotides as soft hybrid nanostructures. J Phys Chem B 2008;112:10942-52. [Crossref] [PubMed]
- Hadorn M, Hotz PE. DNA-mediated self-assembly of artificial vesicles. Plos One 2010;5:e9886 [Crossref] [PubMed]
- Loew M, Kang J, Dähne L, et al. Controlled assembly of vesicle‐based nanocontainers on layer‐by‐layer particles via DNA hybridization. Small 2009;5:320-3. [Crossref] [PubMed]
- Jakobsen U, Simonsen AC, Vogel S. DNA-controlled assembly of soft nanoparticles. J Am Chem Soc 2008;130:10462-3. [Crossref] [PubMed]
- Wang X, Hu J, Liu G, et al. Reversibly switching bilayer permeability and release modules of photochromic polymersomes stabilized by cooperative noncovalent interactions. J Am Chem Soc 2015;137:15262-75. [Crossref] [PubMed]
- Chung M, Koo BJ, Boxer SG. Formation and analysis of topographical domains between lipid membranes tthered by DNA hybrids of different lengths. Faraday Discuss 2013;161:333-45. [Crossref] [PubMed]
- Pei H, Lu N, Wen YL, et al. A DNA nanostructure-based biomolecular probe carrier platform for electrochemical biosensing. Adv Mater 2010;22:4754-8. [Crossref] [PubMed]
- Chan YH, Lenz P, Boxer SG. Kinetics of DNA-mediated docking reactions between vesicles tethered to supported lipid bilayers. Proc Natl Acad Sci U S A 2007;104:18913-8. [Crossref] [PubMed]
- Bridges AA, Zhang H, Mehta SB, et al. Septin assemblies torm by diffusion-driven annealing on membranes. Proc Natl Acad Sci U S A 2014;111:2146-51. [Crossref] [PubMed]
- Cho NJ, Frank CW, Kasemo B, et al. Quartz crystal microbalance with dissipation monitoring of supported lipid bilayers on various substrates. Nat Protoc 2010;5:1096-106. [Crossref] [PubMed]
- Dahlin A, Zäch M, Rindzevicius T, et al. Localized surface plasmon resonance sensing of lipid-membrane-mediated biorecognition events. J Am Chem Soc 2005;127:5043-8. [Crossref] [PubMed]
- Reits EA, Neefjes JJ. From fixed to FRAP: measuring protein mobility and activity in living cells. Nat Cell Biol 2001;3:E145-7. [Crossref] [PubMed]
- Beales PA, Vanderlick TK. DNA as membrane-bound ligand-receptor pairs: cuplex stability is tuned by intermembrane forces. Biophys J 2009;96:1554-65. [Crossref] [PubMed]
- Simons K, Vaz WL. Model systems, lipid rafts, and cell membranes. Annu Rev Biophys Biomol Struct 2004;33:269-95. [Crossref] [PubMed]
- Wen Y, Pei H, Wan Y, et al. DNA nanostructure-decorated surfaces for enhanced aptamer-target binding and electrochemical cocaine sensors. Anal Chem 2011;83:7418-23. [Crossref] [PubMed]
- Chan YH, van Lengerich B, Boxer SG. Effects of linker sequences on vesicle fusion mediated by lipid-anchored DNA oligonucleotides. Proc Natl Acad Sci U S A 2009;106:979-84. [Crossref] [PubMed]
- Pei H, Wan Y, Li J, et al. Regenerable electrochemical immunological sensing at DNA nanostructure-decorated gold surfaces. Chem Commun (Camb) 2011;47:6254-6. [Crossref] [PubMed]
- Gartner ZJ, Bertozzi CR. Programmed assembly of 3-dimensional microtissues with defined cellular connectivity. Proc Natl Acad Sci U S A 2009;106:4606-10. [Crossref] [PubMed]
- Söhl G, Willecke K. Gap junctions and the connexin protein family. Cardiovasc Res 2004;62:228-32. [Crossref] [PubMed]
- Murakami S, Yamaguchi A. Multidrug-exporting secondary transporters. Curr Opin Struct Biol 2003;13:443-52. [Crossref] [PubMed]
- Suntharalingam M, Wente SR. Peering through the pore: nuclear pore complex structure, assembly, and function. Dev Cell 2003;4:775-89. [Crossref] [PubMed]
- Pei H, Liang L, Yao GB, et al. Reconfigurable three-dimensional DNA nanostructures for the construction of intracellular logic sensors. Angew Chem Int Ed Engl 2012;51:9020-4. [Crossref] [PubMed]
- Maingi V, Lelimousin M, Howorka S, et al. Gating-like motions and wall porosity in a DNA nanopore scaffold revealed by molecular simulations. ACS Nano 2015;9:11209-17. [Crossref] [PubMed]
- Yoo JW, Irvine DJ, Discher DE, et al. Bio-inspired, bioengineered and biomimetic drug delivery carriers. Nat Rev Drug Discov 2011;10:521-35. [Crossref] [PubMed]
- Pei H, Li J, Lv M, et al. A graphene-based sensor array for high-precision and adaptive target identification with ensemble aptamers. J Am Chem Soc 2012;134:13843-9. [Crossref] [PubMed]
- McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic Cell membrane remodelling. Nature 2005;438:590-6. [Crossref] [PubMed]
- Yang Y, Wang J, Shigematsu H, et al. Self-assembly of size-controlled liposomes on DNA nanotemplates. Nat Chem 2016;8:476-83. [Crossref] [PubMed]
- Zhang Z, Yang Y, Pincet F, et al. Placing and shaping liposomes with reconfigurable DNA nanocages. Nat Chem 2017;9:653-9. [Crossref] [PubMed]
- Jones MR, Seeman NC, Mirkin CA. Programmable materials and the nature of the DNA bond. Science 2015;347:1260901 [Crossref] [PubMed]
- Dong Y, Sun Y, Wang L, et al. Frame‐guided assembly of vesicles with programmed geometry and dimensions. Angew Chem Int Ed Engl 2014;53:2607-10. [Crossref] [PubMed]
- Douglas SM, Marblestone AH, Teerapittayanon S, et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res 2009;37:5001-6. [Crossref] [PubMed]
- Dietz H, Douglas SM, Shih WM. Folding DNA into twisted and curved nanoscale Shapes. Science 2009;325:725-30. [Crossref] [PubMed]
- Gerling T, Wagenbauer KF, Neuner AM, et al. Dynamic DNA devices and assemblies formed by shape-complementary, non-base pairing 3D components. Science 2015;347:1446-52. [Crossref] [PubMed]
- Marras AE, Zhou L, Su HJ, et al. Programmable motion of DNA origami mechanisms. Proc Natl Acad Sci U S A 2015;112:713-8. [Crossref] [PubMed]
- Howorka S. DNA nanotechnology: bringing lipid bilayers into shape. Nat Chem 2017;9:611-3. [Crossref] [PubMed]
- Schüller VJ, Heidegger S, Sandholzer N, et al. Cellular immunostimulation by CpG-sequence-coated DNA origami structures. ACS Nano 2011;5:9696-702. [Crossref] [PubMed]
- Li J, Pei H, Zhu B, et al. Self-assembled multivalent DNA nanostructures for noninvasive intracellular delivery of immunostimulatory CpG oligonucleotides. ACS Nano 2011;5:8783-9. [Crossref] [PubMed]
- Chen N, Wei M, Sun YH, et al. Self-assembly of Poly-adenine-tailed CpG oligonucleotide-gold nanoparticle nanoconjugates with immunostimulatory activity. Small 2014;10:368-75. [Crossref] [PubMed]
- Li J, Fan C, Pei H, et al. Smart drug delivery nanocarriers with self‐assembled DNA nanostructures. Adv Mater 2013;25:4386-96. [Crossref] [PubMed]
- Söllner TH. Intracellular and viral membrane fusion: a uniting mechanism. Curr Opin Cell Biol 2004;16:429-35. [Crossref] [PubMed]
- Xu Y, Zhang F, Su Z, et al. Hemifusion in SNARE-mediated membrane fusion. Nat Struct Mol Biol 2005;12:417-22. [Crossref] [PubMed]
- Kunishima M, Tokaji M, Matsuoka K, et al. Spontaneous membrane fusion induced by chemical formation of ceramides in a lipid bilayer. J Am Chem Soc 2006;128:14452-3. [Crossref] [PubMed]
- Pei H, Li F, Wan Y, et al. Designed diblock oligonucleotide for the synthesis of spatially isolated and highly hybridizable functionalization of DNA-gold nanoparticle nanoconjugates. J Am Chem Soc 2012;134:11876-9. [Crossref] [PubMed]
- Jahn R, Scheller RH. SNAREs—engines for membrane fusion. Nat Rev Mol Cell Biol 2006;7:631-643. [Crossref] [PubMed]
- You M, Lyu Y, Han D, et al. DNA probes for monitoring dynamic and transient molecular encounters on live cell membranes. Nat Nanotechnol 2017;12:453-9. [Crossref] [PubMed]
- Stengel G, Zahn R, Höök F. DNA-induced programmable fusion of phospholipid vesicles. J Am Chem Soc 2007;129:9584-5. [Crossref] [PubMed]
- Lu N, Pei H, Ge ZL, et al. Charge transport within a three-dimensional DNA nanostructure framework. J Am Chem Soc 2012;134:13148-51. [Crossref] [PubMed]
- Chan YH, van Lengerich B, Boxer SG. Lipid-anchored DNA mediates vesicle fusion as observed by lipid and content mixing. Biointerphases 2008;3:FA17-21. [Crossref] [PubMed]
- Chen X, Araç D, Wang TM, et al. SNARE-mediated lipid mixing depends on the physical state of the vesicles. Biophys J 2006;90:2062-74. [Crossref] [PubMed]
- Kholodenko BN. Cell signalling dynamics in time and space. Nat Rev Mol Cell Biol 2006;7:165-76. [Crossref] [PubMed]
- Wymann MP, Schneiter R. Lipid signalling in disease. Nat Rev Mol Cell Biol 2008;9:162-76. [Crossref] [PubMed]
- Groves JT, Parthasarathy R, Forstner MB. Fluorescence imaging of membrane dynamics. Annu Rev Biomed Eng 2008;10:311-38. [Crossref] [PubMed]
- Schliwa M. Molecular motors. Nature 2003;422:759-65. [Crossref] [PubMed]
- Rudchenko M, Taylor S, Pallavi P, et al. Autonomous molecular cascades for evaluation of cell surfaces. Nat Nanotechnol 2013;8:580-6. [Crossref] [PubMed]
- Zhang DY, Seelig G. Dynamic DNA nanotechnology using strand-displacement reactions. Nat Chem 2011;3:103-13. [Crossref] [PubMed]
- Beales PA. Biophysics: a toehold in cell surface dynamics. Nat Nanotechnol 2017;12:404-6. [Crossref] [PubMed]
- Tkachenko AV. Theory of programmable hierarchic self-assembly. Phys Rev Lett 2011;106:255501 [Crossref] [PubMed]
- Tanner P, Baumann P, Enea R, et al. Polymeric vesicles: from drug carriers to nanoreactors and artificial organelles. Acc Chem Res 2011;44:1039-49. [Crossref] [PubMed]
- Nam J, Beales PA, Vanderlick TK. Giant phospholipid/block copolymer hybrid vesicles: mixing behavior and domain formation. Langmuir 2011;27:1-6. [Crossref] [PubMed]
- Nam J, Vanderlicka TK, Beales PA. Formation and dissolution of phospholipid domains with varying textures in hybrid lipo-polymersomes. Soft Matter 2012;8:7982-8. [Crossref]
Cite this article as: Xiao M, Lai W, Wang X, Qu X, Li L, Pei H. DNA mediated self-assembly of multicellular microtissues. Microphysiol Syst 2018;2:1.


