
Integrating mass spectrometry with microphysiological systems for improved neurochemical studies
Introduction
Until recently, scientists interested in examining cellular-level physiological responses to perturbations have used cultures of cells grown on dishes within a supportive medium. These cell cultures reduce the complexity of the living system to a level at which we can reasonably begin to understand the chemical, physical, and spatial influences affecting cellular behavior. Over the years, reductionist research strategies have advanced a broad range of scientific fields. However, it has become obvious that cells behave differently in cell culture than they do within the body. These differences may lead researchers to incorrect conclusions, with potentially detrimental effects on health-related research and scientific advancement. To solve this issue, and to gain more accurate insight into the physiologically based relationships among cells within the body, there has been a push to develop relevant microsystems in which cells are arranged in layers, cultured with other cells, or seeded onto various structures and geometries or within materials that more closely mimic those found in the body (1-3). The goals are twofold: to improve the relevance of the cell culturing environment through the introduction of chemicals, gradients, cell types, and three-dimensional (3D) structures, and to create platforms that allow spatial and temporal specificity during sampling.
While there are many benefits to using reductionist techniques, they tend to limit the amount of information that can be gained from cells removed from the context in which they develop, especially when investigating dynamic cellular processes. The actual chemical messengers, such as proteins, peptides, and metabolites, rather than genetic differences, are often the markers for change within a biological system. Traditional cell culture systems cannot reproduce the variety of chemical, structural, and mechanical interactions that influence cellular growth, development, communication, and susceptibility to disorders and pathologies (3,4). In fact, cells cultured in 3D systems have been shown to have different morphologies (2,5), biochemical gradients and content (2,5-11), electrophysiological profiles (12), and responses to chemotherapeutics and irradiation (7,8,13), as compared to two-dimensional (2D) cell cultures.
The relevance of the cell culture environment to the physiological system being investigated is particularly important in the study of neurochemistry. The nervous system uses a variety of intercellular messengers, including classical neurotransmitters, neuropeptides, metabolites, lipids, and even gases, to communicate important information throughout the body (14). These intercellular signaling molecules and the information conveyed through them vary depending on their locations, amounts, and degradation pathways within the nervous system (15-17). Additionally, the presence of these molecules and the responses of the target cells also depend on the surrounding environment, changing along with external perturbations (18,19), the presence or absence of other chemical or spatial signals (20,21), and the structural or geometric patterns in a growth system (22). The goal of some neurochemical research is to gain a more complete understanding of the molecules involved in both healthy and diseased neurological systems in order to create better therapeutics for neurological disorders. This is a tall order that involves dynamic monitoring of intracellular, extracellular, signaling, and non-signaling molecules. We need to be able to understand specific relationships between perturbations in the nervous system and the resulting information flow via chemical messengers in both a temporal and spatial manner. While this is a daunting task, the development of in vitro environments that mimic in vivo environments can bring us a step closer to realizing this goal.
In addition to creating physiologically relevant environments through the introduction of chemicals, gradients, and 3D structures or geometries within a system, microphysiological devices also improve the ability to spatially and temporally define physical sampling steps within an experiment (23-28). This is vital to neurochemical investigation in which the specific spatial localization of perturbations allows the study of neural damage and repair, release of cell signaling molecules, and even the specific cells involved in the response. Similarly, temporal relationships between cellular insults and signal transmission are central to gaining an improved understanding of the influence these molecules have on information transfer.
As with advancements in any field, moving toward the development of more physiologically relevant microsystems comes with a need to apply emerging analytical toolsets to the new cell culture platforms and importantly, improve the chemical characterization of microneurological systems. Often, characterization of microphysiological systems is performed using morphological or physiological methods that provide little chemical information. In some cases, immunological techniques are used to target a few preselected molecules for analysis, and/or gene expression profiles determined. However, to be confident that a newly developed system appropriately represents the chemical make-up of the tissue being mimicked, in-depth chemical characterization should be employed. After all, while transcriptomics provides information on the potential of a system, it is the molecules present that dictate its actual physiological state. The improved physiological relevance of microphysiological systems should lead to important new discoveries, particularly in the neurochemical or cell signaling realm; therefore, we should be focusing on ways to better interrogate the systems for unique chemical information. Mass spectrometry (MS) is the single most information-rich chemical characterization technique available, allowing the detection and identification of a wide range of molecules in both a targeted and an untargeted manner. We expect that through the coupling of MS to these microsystems, we will be able to better characterize the dynamic chemical make-up of the cells involved, as well as the molecular messengers used in the information flow between cellular groups, both under physiological conditions and as a result of external perturbations.
For those of us who are more accustomed to thinking about reductionist approaches, there are multiple examples of biologists and engineers working together to create innovative platforms for the development of relevant microsystems. In this review, we broadly outline MS approaches of relevance to these systems, discuss the use of microfluidic devices, spheroids, hydrogels, and scaffolds to create appropriate microsystems, and lastly, highlight the exciting progress and future potential of interfacing these approaches to obtain improved chemical information for advancing neurochemical research.
MS as a chemical characterization approach
Introduction to MS
While the goal of many microsystem designs is to create an improved cell culturing environment and enhanced sampling system defined by the spatial and temporal aspects of the device, eventually the samples need to be characterized. If a preselected set of molecules are of interest, specific molecular or affinity probes can be used. However, for more untargeted or less well-characterized analytes, MS remains the most chemically information-rich approach because it allows the characterization and quantitation of thousands of molecular species, usually without analyte preselection. There are a multitude of modern mass spectrometric techniques to choose from. The method selected depends on several factors, including sample properties and instrumental parameters, which together determine the compounds detected. Although detailed descriptions of all of the available hardware and operating procedures for MS are not within the scope of this review, a brief overview is provided to aid in the selection of the appropriate MS-based characterization approach. Critical considerations include choosing the appropriate sample conditioning method (including a separation step), vaporization and ionization approaches, and mass analyzer. Readers interested in learning more about MS are referred to several recent reviews (29-31).
Analysis of small sample volumes via MS
Researchers interested in characterizing a wide range of molecules using a microphysiological system, either present in the environment surrounding the system or found within the system, can efficiently couple the device to a mass spectrometer for the collection of small-volume samples. The most straightforward microphysiological systems for this type of sampling are those with direct inlet and outlet ports and/or direct access to the material that will be collected. The next steps depend on the desired information and the MS instrumentation available. The two most commonly employed MS characterization approaches are matrix-assisted laser desorption/ionization (MALDI) and electrospray ionization (ESI), as they are both well suited for small-volume samples (32-36).
For a dried sample, MALDI MS is a fast method that involves incorporating the analyte with an organic matrix, which when irradiated with a laser, vaporizes and ionizes the sample (34,37). MALDI measurements have been performed from cellular releasates, individual cells, and even directly from tissue slices (38). Basically, if a small-volume (picoliter to microliter) sample is dried and the appropriate chemical matrix added, efficient detection of a broad range of molecules becomes possible. MALDI has a high salt tolerance and requires nanoliter-volume samples (27,32,39), making it useful for the analysis of small-volume biological samples, and even individual cells (40-42). Additionally, for precious samples, the recovery of leftover analytes is possible (43). Sample conditioning can enhance MALDI analysis, for example, by desalting and concentration via solid phase extraction (SPE) (44,45). After extraction, analytes of interest can be eluted onto a target plate (by SPE) or dried down, reconstituted in an appropriate amount of solvent, and then placed on the plate (by SPE or liquid extraction). In both situations, the sample is plated, mixed with a matrix, and dried in preparation for mass spectrometric analysis.
While MALDI has a number of advantages, an issue with MALDI is that interfacing it to separations, such as liquid chromatography (LC) or capillary electrophoresis (CE), adds complexity as compared to ionization techniques that use samples in solution. Without a preseparation with MALDI, ion suppression can occur; thus, there are applications that are better suited to LC-ESI-MS. In ESI-MS, a liquid sample is sprayed from a capillary via the application of an electrical potential to the end of the capillary (35,46). The electrosprayed droplets are desolvated and vaporized, resulting in multiply charged ions, which are introduced into the mass analyzer (35). ESI works well as a characterization approach after a liquid phase separation, and so often follows CE (44,47) or LC (48). The figures of merit of ESI make it particularly useful for the identification and quantitation of a wide range of analytes within complex samples (35). Researchers interested in evaluating the analytes within a microphysiological system would benefit from using these techniques, although other MS ionization approaches are available (49).
Analysis of large, complex samples via MS
At the other end of the volume scale, a number of metabolomics or proteomics approaches have evolved that take larger-volume samples, condition them, and then perform an LC separation followed by high performance MS, including tandem MS (MS/MS) (48). As mentioned in the previous section, LC separation is important for the reduction of the chemical complexity and dynamic range inherent to biological samples, with the goal of separating the samples into chemically simpler fractions. LC-MS/MS generally requires larger amounts of sample than direct MALDI MS profiling, but is useful for peptidomic, metabolomic, or proteomic studies, as well as for quantitation.
Direct imaging of samples using MS
MALDI MS is increasingly being employed for spatially resolved tissue characterization in an approach termed mass spectrometry imaging (MSI). In a common embodiment of MSI, a laser samples the surface in a raster pattern, and at each point, a mass spectrum is acquired. The resulting spectra are then used to create images of ion intensity at specific locations within the sample (50). Similar to interrogating a tissue with MS, one can use MSI to spatially measure the compounds within a microfabricated device (51,52). Additionally, because microphysiological systems can contain complex 3D structures, these structures can be removed from the systems and processed as though they are tissue through a series of stabilization and sectioning steps prior to analysis (38,53). MSI can be performed using MALDI MS, secondary ion MS (SIMS), or desorption ESI MS (54). Each ionization method has distinct figures of merit as well as different sample preparation requirements (50,54). No matter which detection modality is used, MSI is useful for researchers who want to obtain spatial information on the molecular content of their samples, both for basic study and to determine how well the model systems compare to the in vivo systems they are mimicking.
Single cell measurements via MS
Understanding the chemical differences between cells is an important objective for neurochemical studies. Various MS methods can be employed for single cell measurements, including the aforementioned MALDI, ESI, and SIMS (38). Single cells contain femtoliters to picoliters of liquid and, therefore, measurements require high sensitivity and low limits of detection. Additionally, cells contain a complex mixture of molecules, requiring wide analyte coverage and ionization of intact biomolecules. Direct sampling from specific cells using liquid microjunction extraction for CE-ESI MS (55), and direct MALDI profiling of individual cells sorted into microarrays (42) or dissociated onto slides (56), are methods that are currently employed to interrogate the chemical content of single cells, offering great potential for integration with microphysiological systems.
Sample preparation considerations for MS
Here we outline the factors that impact the selection of the sampling and measurement protocols. For sampling from a microphysiological system, we expect that cells, tissues, and extracellular fluid would be the most common types of samples obtained. The sample preparation method selected for an MS analysis depends on a variety of factors, including the tissue or model the sample came from, the chemical information the researcher is trying to obtain, the instrument that will be used for the analysis, and how much time will elapse between sample preparation and analysis. In addition, a decision has to be made about whether it is important to the experimental objective to maintain structural, temporal, or location information, a choice that impacts how samples are collected and prepared.
For example, if chemical characterization of different cellular populations is the goal, cells could be detached (or “dissected”) from the device, suspended in a liquid media, stabilized with glycerol or a similar substance so that they remain intact during a drying step, placed on a slide or MALDI target, washed to remove the excess glycerol, dried under a nitrogen stream, and coated with a matrix in preparation for MALDI. Alternatively, the cells could be individually prepared for intracellular media sampling by CE. These sampling approaches would allow for intracellular chemical characterization of different cell types, but would not maintain the spatial relationships of the cells. In studies where retaining spatially relevant chemical information is more important, the system could instead be frozen, cryogenically sliced, and then the slices deposited onto MALDI target plates, stabilized, and covered in a matrix for MS analysis. This type of sample preparation maintains intra- and extracellular areas as well as spatial relationships between cells. On the other hand, if analysis of cell signaling molecules is the goal, extracellular fluid can be collected from the device in a temporal or spatially relevant manner, subjected to sample clean-up via SPE to remove the salts from the extracellular media, and then either mixed with a MALDI matrix or prepared with an appropriate solvent for LC-MS analysis. Finally, if the goal is to understand as much chemical information as possible about a certain system (at the expense of spatial detail), the cells or tissues can be homogenized, the proteins, peptides, small molecules, or metabolites extracted, and the sample subjected to an SPE step and then prepared for LC prior to MS analysis. These are just a few examples of sample preparation methods that can be employed prior to MS analysis.
Microphysiological systems for neurobiological studies
Microfluidic systems
Microfluidic systems for biological applications rely on the physics and chemistry of small-volume materials and fluidic interactions to create intricate platforms for the growth, analysis, and manipulation of cells on a micrometer scale (57). When applied to biological systems, they have historically been used as a reductionist approach for single cell analyses, cell sorting, and compartmentalization; however, microfluidic devices are also used to create microphysiological systems (28,58-63). The small-volume fluidic interactions allow for the formation of specially controlled microenvironments that can mimic the physiological milieu and structural relationships, while also providing opportunities for sampling, observation, and introduction of specific perturbations (64,65). These attributes allow for the design of studies to provide a more accurate picture of cellular responses. Our research group has had success using microfluidic devices to sample from cells for mass spectrometric analysis (27,51,52,66), and we look forward to performing similar experiments utilizing microphysiological systems.
The move away from traditional cell culture and toward constructing a microphysiological system can be facilitated by co-culturing different cell types in a microfluidic device. While many fields use the technology, the intricate relationships between neurons and their surrounding cells make co-culture for neurochemical investigation particularly relevant. First and foremost, neurons and glia are interdependent, and studies have shown that neurons behave differently with and without glia in their cultures (28,58,61). Therefore, both types of cells need to be incorporated into the microfluidic device. For example, Majumdar et al. (58) demonstrated a microfluidic device for the co-culture of primary hippocampal neurons and glial cells in which both types of cells were not physically in contact, but glial-conditioned media flowed directly from glia to the neurons. They found that the neurons co-cultured with glia had greater transfection efficiency than traditional cell cultures, demonstrating the influence of glial presence in the culture system. Li and Ren et al. (28) developed a microfluidic device in which axonal injury can be studied within a system that includes both neurons and glia. They showed that their system allowed re-growth of axons when glial cells were introduced to the culture system. With the understanding that neuronal/glial interactions are important to study, Park et al. (59) kept the neuronal cell bodies physically separated, but allowed interaction between oligodendrocytes and the neuronal axons for the study of myelination (Figure 1A).
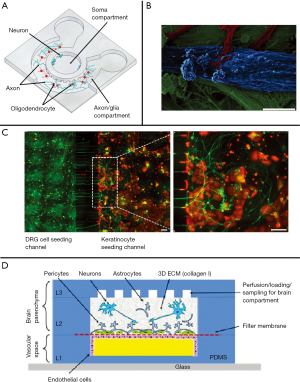
With microphysiological systems, it is important that target tissues are incorporated within the model system. Neurons develop, regenerate, and function based on their surroundings (14,28,69), supporting the need for functional neuromuscular junctions, sensory systems, and brain structures that can be studied. The neuromuscular junction consists of the spinal motor neurons, the cell bodies of which can be found in the ventral horn of the spinal cord, and their target organ, muscle. Recently, Kim et al. (60) demonstrated a microsystem in which spinal cord and muscle tissues were placed within channels located on opposing sides of the device, connected with microchannels. Axons from the spinal cord grew across the microchannels and innervated the muscle tissue. Southam et al. (61) described the use of a commercial dual-chamber microfluidic device to culture motor neurons and glia on one side of the device and myocytes on the other. They demonstrated the growth of motor neuron axons through the microchannels and into the myotube side (Figure 1B), and discussed the importance of glial cells and target muscle tissue for the health of the motor neuron in culture. Perhaps more exciting is a system reported by the Perlson group (20,62), created using a slice of an embryonic spinal cord explant co-cultured with myotubes. In this microfluidic device, the axons from the motor neurons grew through grooves to innervate the myotubes. One of their studies revealed not only that the axons grew more quickly through the grooves when the myoblasts were present than when they are not, but also that the growth factor glial cell line-derived neurotrophic factor only promoted axonal growth and innervation when applied to the axons of the motor neurons, and not when applied to the cell body (20). This outcome is a demonstration of the important new biochemical insights that can be gained from using microphysiological systems.
For the peripheral sensory nervous system, there is one more “side” to account for within the microsystem. The cell body of the sensory neuron resides in the sensory ganglia located near the spinal cord (70,71). Sensory neurons only have one outgrowth, an axon, which bifurcates, extending one branch to the skin or viscera and the other into the dorsal horn of the spinal cord. Using a popular microfluidic device design (72), Tsantoulas and colleagues (67) created the sensory ending side of this system by culturing primary sensory neurons from the dorsal root ganglion on one side of a two-chamber device and keratinocytes on the other, to resemble the skin (Figure 1C). They demonstrated the successful innervation of the keratinocytes by the sensory neurons, as have other groups using separate two-compartment co-culture models (21,73,74). As for the side of the sensory neuron axon that extends to the spinal cord, a few groups have demonstrated functional cultures between neurons of the dorsal horn of the spinal cord and those of the dorsal root ganglia (75-78). Additionally, Johnson et al. (26) demonstrated a 3D printed tri-chamber device within which they successfully cultured peripheral sensory neurons, Schwann cells and either epithelial cells or hippocampal neurons in separate chambers. While hippocampal neurons are not secondary sensory neurons, this is one of the only successful central nervous system (CNS) to peripheral nervous system micro-cultures. Unfortunately, there are not yet any microsystems that can mimic the peripheral sensory system from skin to spinal cord; however, work is being done in this area.
Another important area of research for the design of microfluidic systems includes the re-creation of the blood-brain barrier (BBB). Understanding the BBB is important to the study of neurochemistry because it is the gatekeeper of the brain, regulating which molecules are allowed to enter the CNS from the blood. Various groups have designed devices that mimic certain aspects of the BBB (79). Two similar device designs, one created by the Sundaram group (63) and another by the Wikswo group (68), successfully modeled the neurovascular unit by using multiple layers, including a neural chamber and a vascular chamber, and a microporous membrane separating the two (Figure 1D) (68). The neural chamber consisted of a physiologically relevant mix of neurons and supported cells such as astrocytes and microglia (for the Sundaram group) or pericytes (for the Wikswo group), and the vascular chamber contained microvascular endothelial cells. Both groups reported the ability to include flow within their design, which is imperative for the appropriate function of endothelial cells and for mimicking vasculature. Importantly, the Wikswo group seeded the neurons and astrocytes within a collagen gel in the “brain” chamber to even better model brain structure, and also used human-derived cells within their system. Another noteworthy BBB device design is the Ingber group’s (80) use of a microfluidic system to create a cylindrical lumen inside a microchannel within which astrocytes, pericytes, and endothelial cells were seeded. This work is important because it incorporates the appropriate shape of the blood vessels (cylinder), unlike the rectangular channels used by other groups, and also removes the membrane barrier between cell types, allowing them to be in physical contact with each other, as they are in the body.
Spheroids
Another way to create physiologically relevant nervous system models is to place neurons and their support cells together and let them assemble or aggregate on their own. The creation of spheroid cultures relies on the cells’ natural proclivity to aggregate, rather than on the strict design of structural, chemical, and physical relationships used by microfluidic devices. A spheroid culture is both a way to create a more natural state for the cells under investigation, and a means to study the natural formation of cellular aggregates. Spheroid culturing technique is based on keeping cells in suspension so that they will join together, rather than settle on and form attachments to a particular cell-culturing surface. Aggregation can be encouraged through stirring, suspending cells in drops from a surface, and plating in non-adhesive microarray wells. Once aggregated, the spheroids tend to demonstrate behavior similar to that found in vivo, such as exhibiting the proper intercellular relationships or secreting their own extracellular matrix (ECM) (81). The creation of spheroids and spheroid networks can be useful in the study of intercellular interactions and cell-ECM interactions (81), toxicity studies (82), and even neural tissue transplantation (83).
As an example of spheroid neural cultures being used to study disease, the Lee lab (82) built an in vitro model of Alzheimer’s disease using a network of neurospheres to model the layered cellular architecture of the brain’s cortex for the study of beta amyloid exposure. Building on this model, Park et al. (84) incorporated the constant fluid flow that is found in physiological systems. The potential implications for their model in the study of Alzheimer’s disease is important to note, particularly for understanding the response of the tissue to beta amyloid exposure as well as for toxicity studies for potential therapeutics.
Another group, led by Kato-Negishi (83), created a network of neurospheres that could then be used as a stamp and placed directly onto cortical tissue. While it is unclear how this type of brain tissue would actually function once incorporated into a living system, the researchers did show integration between the stamped neurosphere network and the whole brain tissue, which is a promising step forward for future cognitive interface work.
Recently, Pamies et al. (85) demonstrated the creation of a reproducible, size-controlled spheroid culture using induced pluripotent stem cells differentiated into a structure containing appropriately localized neurons, oligodendrocytes, and astrocytes as well as neural sub-populations, such as those expressing dopamine, glutamine, or GABA. Since they are derived from human cells and are uniformly made, these brain microphysiological systems, as they are called, could be particularly useful for pharmaceutical or neurotoxicity studies.
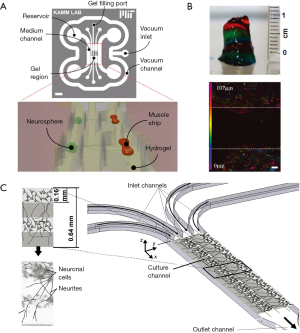
Spheroids can also be used within microfluidic devices. For example, the Kamm group (65) described an enhanced co-culture microfluidic device in which a 3D system was created that allows for the culture of both a neurosphere on one side of the device and a muscle strip on the other, with axonal outgrowth from the neurosphere connecting to the muscle strip (Figure 2A).
Hydrogels, fibers, and other soft materials
The incorporation of various soft materials into cell culture is another way that physiological relevance can be enhanced in an engineered system. Hydrogels are highly absorbent cross-linked polymers that can be used in a variety of ways. They are particularly useful in the creation of microphysiological systems because they are mechanically and structurally similar to tissue and ECM (88). This similarity to natural tissue, combined with a level of porosity that allows for cell migration and nutrient and waste exchange, make hydrogels prime materials for creating 3D cell cultures (86,88).
Lozano et al. (86) designed a system in which a peptide-modified polymer, Gellan gum-RGD, was mixed with cortical cells and printed using a 3D printing system into a layered structure similar to that of the cortex (Figure 2B). The authors found that the neurons and glia were functional, and that synapses formed between printed layers. Therefore, they succeeded in creating a 3D cortex-like structure with viable neurons encapsulated inside, demonstrating appropriate growth and neurite extension. Similarly, Kunze et al. (87) used microfluidic devices to create alginate-enriched agarose hydrogel layers that model the structure of the cortex (Figure 2C). The layers can also be perfused with nutrients and/or stimulated with chemicals in a gradient and are oriented in the x-y plane for facile microscope imaging. While their system used only one type of cell, the importance of their device in correctly simulating the external environment for the neurons of the cortex cannot be dismissed.
Huval et al. (89) created a 3D system in which a dorsal root ganglion explant was placed within a dual hydrogel encasement system that had been treated with both growth-permissive and growth-inhibiting molecules so that the axonal outgrowth followed a discrete nerve-like path. This “microscale organotypic model of peripheral nerve tissue,” as they described it, has a similar structure and electrophysiological function as nerve tissue, making it a potential model for use in pharmaceutical and clinical testing. It would also be an interesting model for injury, regeneration, and neurochemical studies.
A multi-layered microfluidic device to fabricate hydrogel constructs that mimic the structure of a nerve bundle, with its complex cross-sectional morphology, was designed by Kitagawa et al. (90). They used sodium alginate to create stiff microfibers that were encapsulated by a softer hydrogel, which included sodium alginate mixed with propylene glycol alginate. The PC12 neuron-like cells grew along the stiff microfibers, creating an analogous structure to an axon surrounded by Schwann cells.
Hydrogels influence cellular responses based on the stiffness, charge, and other properties of the gel (91). While the ability to tune a hydrogel creates unlimited opportunities for creating various environments within which cells can grow, the sensitivity of the cells to the hydrogel environment may not make them optimal for the study of in vivo responses, unless a hydrogel that has the exact same parameters as the cell’s environment is used.
Daud et al. (92) created nerve-like structures via electrospun polycaprolactone fiber scaffolds. These fibers, aligned in groups and created with uniform diameters of 1, 5, or 8 µm, allowed for the alignment of neurons, axons, and Schwann cells as they grew in culture. They used primary cell cultures and dorsal root ganglion explants, observing that neuronal and Schwann cell co-localization occurred for the dorsal root ganglion cultures, but not as extensively for the neuronal and primary Schwann cell cultures. The authors also studied primary neuron cell culture versus neuron/Schwann cell co-culture and concluded that axons extended further in the presence of Schwann cells than without. Both results add evidence to the importance of using an in vitro system that is the most closely related to the in vivo system.
A microscaffold system made from an array of microfabricated towers is the solution that Rowe et al. (93) designed to overcome the lack of appropriate circulation to 3D cell cultures. They used hollow microtowers that contained multiple fluid ports, which allowed for media and nutrient perfusion throughout the device. This group also included electrodes on the towers for ease of future electrophysiological stimulation or recording.
As a final example, Zou et al. (94) used self-assembly of the peptide IKVAV to create a scaffold of nanofibers, which were useful for guiding the outgrowth of axons from a dorsal root ganglion explant.
Applying MS to microphysiological systems for chemical analyses
The microsystems highlighted in this review were initially developed to improve scientists’ ability to model in vivo physiology, with many used to perform morphological, immunofluorescent, and even some biochemical analyses. Each of the microfluidic systems discussed are also well suited for chemical integration with mass spectrometric analysis and can be used to collect cellular releasates, or would only need small adjustments in the form of stimulation and sampling ports for coupling with off-chip analysis by MS.
Microfluidic devices have a history of being coupled with mass spectrometric analysis (27,52,95-98), and we expect this will continue. Perfusion of media through input and output ports for stimulations and sample collection is the simplest approach, while others might opt for employing microdialysis-like methods, or adding more intricate on-chip processing steps to the sample collection (96,98). Although these adjustments could require changes to the design and fabrication of a microfluidic device, existing media perfusion ports can also be used for perturbations or sampling. For designs that do not have direct access, holes of varying diameters can be punched into the devices using biopsy punches or via less invasive sampling methods, such as a syringe and needle. Finally, researchers may opt to revise their designs and fabrication methods to create an optimized interface between their devices and the mass spectrometer.
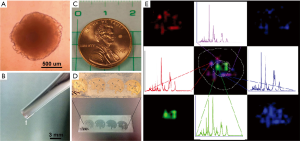
Neurospheres can be incorporated into MS in two ways. First, they can be cultured within a microfluidic framework, as described by Uzel et al. (65). Within a microfluidic device, neurospheres would provide a physiologically relevant cell culture construct and sampling would occur through perfusion, similarly to how it is outlined above. Additionally, the cellular and chemical composition of a neurosphere, as well as its ability to create its own ECM, would be interesting to study using MSI. The neurospheres can be sectioned and then subjected to MSI to examine their chemical make-up, as described by the Hummon group (53,54) in their study of spherical cultures (Figure 3) (54). Impressively, they were able to identify proteins and their distributions within colon carcinoma spheroids. Additionally, the surroundings of the neurospheres could be directly imaged using SIMS or MALDI without sectioning, helping to better understand the development of the ECM and surrounding environment, similar to how our group images biofilms (99).
Systems created out of hydrogels, scaffolds, and fibers can be sectioned and imaged similarly to neurospheres, or can be perfused and have analytes collected via sampling from device output ports, similar to sampling from microfluidic devices. Additionally, procedures such as microdialysis may be successful for sampling from within a hydrogel or scaffolded “tissue”. Specifically for polymer-based systems, one issue that may occur is that even if sampling is successful, the polymer content may suppress or overwhelm the signal from the sample. However, the current success in coupling MS with microfluidic devices that employ polymer materials, as described above, the use of hydrogels for localized protein extraction prior to MS analysis (100), and the compatibility of MS with microsystems using poly(dimethylsiloxane) (101), polycarbonate (102), and poly(methyl methacrylate) (103), demonstrate that polymer contamination from devices may not be as large an issue as some have assumed. Should concerns arise when using specific polymer-based devices, these issues could be potentially resolved through data analysis techniques, such as subtraction of the peaks associated with the intervening polymers. Further, the development of non-polymer-based materials, improved sample clean-up techniques, and increased instrumental resolution and sensitivity could be future solutions to potential polymer contamination issues.
Additionally, single cell measurements for the detection of rare cells or the measurement of appropriate cell ratios/types within the system can be performed through direct sampling from the microphysiological device or by harvesting the cells through enzymatic digestion and dissociation and measuring their cell profiles using microarrays (42) or microscopy-guided MS analysis after dispersion (56).
We acknowledge that a limitation of using MS to chemically characterize microphysiological systems is the potentially low amount of analyte present in such small systems, particularly when performing non-targeted analyses. However, recent improvements to mass analyzers and sample introduction systems have led to large decreases in the sample amounts required, with work using zeptomole amounts of metabolites and proteins now being reported (104-106). However, when using MS for these small-volume studies, researchers are cautioned that although the chemical information obtained by MS analysis does not require analyte preselection, the resulting data is often not comprehensive.
Finally, when working with human tissue samples, there are often time delays, and issues with sampling variability and non-uniform storage conditions, all of which can greatly impact the chemical information obtained from the tissues. When robust microphysiological systems become available, they will be an excellent source of samples from which we could gain a multitude of chemical details about normal human growth, development, and aging, as well as disease progression. It is important to develop the analytical techniques now for when that time comes.
Conclusions
For years, the study of neurochemistry has relied on both in vivo systems, which can be difficult to control, and a range of in vitro 2D cell cultures and organotypic slices, which offer improved control and access to the cells but are not ideal models for living systems. Now, thanks to both improved microtechnologies and a better understanding of the nervous system, we have the tools at hand to build in vitro systems that behave more closely to in vivo systems.
A range of MS measurement approaches offer the most information-rich, non-targeted chemical characterization options available for characterizing metabolites, peptides, and proteins within a living system. Coupling microphysiological systems to mass spectrometers requires attention to the hyphenation details in order to obtain the greatest information on a dynamic cellular environment without introducing biases. While the analytical performance for mass spectrometers is impressive, there are numerous opportunities for improved interfaces between microphysiological systems and mass spectrometric measurement technologies, and we expect systems with enhanced spatial, chemical, and temporal resolution to be introduced in the coming years. Presently there are only a few examples of microphysiological systems having been used for analytical inquiries, and even fewer in neurochemistry; nonetheless, we look forward to the neurochemical insights to be gained from continued research in this area. We envision exciting progress in the integration of microphysiological systems with mass spectrometric analysis, and expect that the high information content of MS will enable new discoveries about the molecular players in both health and disease.
Acknowledgments
Funding: This work was supported by the National Institute on Drug Abuse under Award No. P30 DA018310, and the National Science Foundation under Award Nos. CHE-16-06791 and DGE-17-35252.
Footnote
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/mps.2018.05.01). Emily G. Tillmaand and Jonathan V. Sweedler report grants from National Institute of Drug Abuse, grants from National Science Foundation, during the conduct of the study. The authors have no other conflicts of interest to declare.
Disclaimer: The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wikswo JP. The relevance and potential roles of microphysiological systems in biology and medicine. Exp Biol Med (Maywood) 2014;239:1061-72. [Crossref] [PubMed]
- Baker BM, Chen CS. Deconstructing the third dimension: how 3D culture microenvironments alter cellular cues. J Cell Sci 2012;125:3015-24. [Crossref] [PubMed]
- Huang S, Wikswo J. Dimensions of Systems Biology. Reviews of Physiology Biochemistry and Pharmacology. Berlin, Heidelberg: Springer, 2007:81-104.
- Li XJ, Valadez AV, Zuo P, et al. Microfluidic 3D cell culture: potential application for tissue-based bioassays. Bioanalysis 2012;4:1509-25. [Crossref] [PubMed]
- Baharvand H, Hashemi SM, Kazemi Ashtiani S, et al. Differentiation of human embryonic stem cells into hepatocytes in 2D and 3D culture systems in vitro. Int J Dev Biol 2006;50:645-52. [Crossref] [PubMed]
- Yue X, Lukowski JK, Weaver EM, et al. Quantitative Proteomic and Phosphoproteomic Comparison of 2D and 3D Colon Cancer Cell Culture Models. J Proteome Res 2016;15:4265-76. [Crossref] [PubMed]
- Kumar HR, Zhong X, Hoelz DJ, et al. Three-dimensional neuroblastoma cell culture: proteomic analysis between monolayer and multicellular tumor spheroids. Pediatr Surg Int 2008;24:1229-34. [Crossref] [PubMed]
- Desoize B, Jardillier J. Multicellular resistance: a paradigm for clinical resistance? Crit Rev Oncol Hematol 2000;36:193-207. [Crossref] [PubMed]
- Edmondson R, Broglie JJ, Adcock AF, et al. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev Technol 2014;12:207-18. [Crossref] [PubMed]
- Benya PD, Shaffer JD. Dedifferentiated chondrocytes reexpress the differentiated collagen phenotype when cultured in agarose gels. Cell 1982;30:215-24. [Crossref] [PubMed]
- Zietarska M, Maugard CM, Filali-Mouhim A, et al. Molecular description of a 3D in vitro model for the study of epithelial ovarian cancer (EOC). Mol Carcinog 2007;46:872-85. [Crossref] [PubMed]
- Frega M, Tedesco M, Massobrio P, et al. Network dynamics of 3D engineered neuronal cultures: a new experimental model for in-vitro electrophysiology. Sci Rep 2014;4:5489. [Crossref] [PubMed]
- Shield K, Ackland ML, Ahmed N, et al. Multicellular spheroids in ovarian cancer metastases: Biology and pathology. Gynecol Oncol 2009;113:143-8. [Crossref] [PubMed]
- Squire L, Berg D, Bloom F, et al., editors. Fundamental Neuroscience. Third ed.: Elsevier, 2008.
- Strand FL, editor. Neuropeptides: Regulators of Physiological Processes. Cambridge, MA: The MIT Press, 1998.
- Xu XJ, Wiesenfleld-Hallin Z. Neuropeptides: Pain. In: Squire LR, editor. Encyclopedia of Neuroscience. Oxford: Academic Press, 2009:931-4.
- Parenti C, Aricò G, Ronsisvalle G, et al. Supraspinal injection of Substance P attenuates allodynia and hyperalgesia in a rat model of inflammatory pain. Peptides. 2012;34:412-8. [Crossref] [PubMed]
- Hubbard RD, Quinn KP, Martínez JJ, et al. The role of graded nerve root compression on axonal damage, neuropeptide changes, and pain-related behaviors. Stapp Car Crash J 2008;52:33-58. [PubMed]
- Lee KE, Winkelstein BA. Joint distraction magnitude is associated with different behavioral outcomes and substance P levels for cervical facet joint loading in the rat. J Pain 2009;10:436-45. [Crossref] [PubMed]
- Zahavi EE, Ionescu A, Gluska S, et al. A compartmentalized microfluidic neuromuscular co-culture system reveals spatial aspects of GDNF functions. J Cell Sci 2015;128:1241-52. [Crossref] [PubMed]
- Roggenkamp D, Falkner S, Stäb F, et al. Atopic keratinocytes induce increased neurite outgrowth in a coculture model of porcine dorsal root ganglia neurons and human skin cells. J Invest Dermatol 2012;132:1892-900. [Crossref] [PubMed]
- Millet LJ, Gillette MU. New perspectives on neuronal development via microfluidic environments. Trends Neurosci 2012;35:752-61. [Crossref] [PubMed]
- Tsamandouras N, Chen WLK, Edington CD, et al. Integrated Gut and Liver Microphysiological Systems for Quantitative In Vitro Pharmacokinetic Studies. AAPS J 2017;19:1499-512. [Crossref] [PubMed]
- Blundell C, Tess ER, Schanzer AS, et al. A microphysiological model of the human placental barrier. Lab Chip 2016;16:3065-73. [Crossref] [PubMed]
- Kilic O, Pamies D, Lavell E, et al. Brain-on-a-chip model enables analysis of human neuronal differentiation and chemotaxis. Lab Chip 2016;16:4152-62. [Crossref] [PubMed]
- Johnson BN, Lancaster KZ, Hogue IB, et al. 3D printed nervous system on a chip. Lab Chip 2016;16:1393-400. Erratum in: Lab Chip 2016;16:1946. [Crossref] [PubMed]
- Croushore CA, Supharoek SA, Lee CY, et al. Microfluidic device for the selective chemical stimulation of neurons and characterization of peptide release with mass spectrometry. Anal Chem 2012;84:9446-52. [Crossref] [PubMed]
- Li L, Ren L, Liu W, et al. Spatiotemporally controlled and multifactor involved assay of neuronal compartment regeneration after chemical injury in an integrated microfluidics. Anal Chem 2012;84:6444-53. [Crossref] [PubMed]
- Glish GL, Vachet RW. The basics of mass spectrometry in the twenty-first century. Nat Rev Drug Discov 2003;2:140-50. [Crossref] [PubMed]
- Maher S, Jjunju FPM, Taylor S. Colloquium: 100 Years of Mass Spectrometry: Perspectives and Future Trends. Rev Mod Phys 2015;87:113-35. [Crossref]
- Rubakhin SS, Sweedler JV. A mass spectrometry primer for mass spectrometry imaging. Methods Mol Biol 2010;656:21-49. [Crossref] [PubMed]
- Romanova EV, Aerts JT, Croushore CA, et al. Small-volume analysis of cell-cell signaling molecules in the brain. Neuropsychopharmacology 2014;39:50-64. [Crossref] [PubMed]
- Hillenkamp F, Karas M. The MALDI Process and Method. MALDI MS. Wiley-VCH Verlag GmbH & Co. KGaA, 2007:1-28.
- Dreisewerd K. Recent methodological advances in MALDI mass spectrometry. Anal Bioanal Chem 2014;406:2261-78. [Crossref] [PubMed]
- Wilm M. Principles of Electrospray Ionization. Mol Cell Proteomics 2011;10:M111.009407.
- Qi M, Philip MC, Yang N, et al. Single Cell Neurometabolomics. ACS Chem Neurosci 2018;9:40-50. [Crossref] [PubMed]
- Watson JT, Sparkman OD. Introduction to Mass Spectrometry: Instrumentation, Applications, and Strategies for Data Interpretation. Wiley, 2013.
- Comi TJ, Do TD, Rubakhin SS, et al. Categorizing Cells on the Basis of their Chemical Profiles: Progress in Single-Cell Mass Spectrometry. J Am Chem Soc 2017;139:3920-9. [Crossref] [PubMed]
- Signor L, Boeri Erba E. Matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) mass spectrometric analysis of intact proteins larger than 100 kDa. J Vis Exp 2013;(79).
- Jansson ET, Comi TJ, Rubakhin SS, et al. Single Cell Peptide Heterogeneity of Rat Islets of Langerhans. ACS Chem Biol 2016;11:2588-95. [Crossref] [PubMed]
- Romanova EV, Rubakhin SS, Monroe EB, et al. Single Cell Mass Spectrometry. Single Cell Analysis. Wiley-VCH Verlag GmbH & Co. KGaA, 2009:109-33.
- Urban PL, Jefimovs K, Amantonico A, et al. High-density micro-arrays for mass spectrometry. Lab Chip 2010;10:3206-9. [Crossref] [PubMed]
- Cockrill SL, Foster KL, Wildsmith J, et al. Efficient micro-recovery and guanidination of peptides directly from MALDI target spots. Biotechniques 2005;38:301-4. [Crossref] [PubMed]
- Ong TH, Tillmaand EG, Makurath M, et al. Mass spectrometry-based characterization of endogenous peptides and metabolites in small volume samples. Biochim Biophys Acta 2015;1854:732-40.
- Hatcher NG, Richmond TA, Rubakhin SS, et al. monitoring activity-dependent peptide release from the CNS using single-bead solid-phase extraction and MALDI TOF MS detection. Anal Chem 2005;77:1580-7. [Crossref] [PubMed]
- Fenn JB, Mann M, Meng CK, et al. Electrospray Ionization–Principles and Practice. Mass Spectrom Rev 1990;9:37-70. [Crossref]
- Klepárník K. Recent advances in the combination of capillary electrophoresis with mass spectrometry: from element to single-cell analysis. Electrophoresis 2013;34:70-85. [Crossref] [PubMed]
- Niessen WM. Progress in liquid chromatography-mass spectrometry instrumentation and its impact on high-throughput screening. J Chromatogr A 2003;1000:413-36. [Crossref] [PubMed]
- Wu C, Dill AL, Eberlin LS, et al. Mass spectrometry imaging under ambient conditions. Mass Spectrom Rev 2013;32:218-43. [Crossref] [PubMed]
- Shariatgorji M, Svenningsson P, Andrén PE. Mass spectrometry imaging, an emerging technology in neuropsychopharmacology. Neuropsychopharmacology 2014;39:34-49. [Crossref] [PubMed]
- Jo K, Heien ML, Thompson LB, et al. Mass spectrometric imaging of peptide release from neuronal cells within microfluidic devices. Lab Chip 2007;7:1454-60. [Crossref] [PubMed]
- Zhong M, Lee CY, Croushore CA, et al. Label-free quantitation of peptide release from neurons in a microfluidic device with mass spectrometry imaging. Lab Chip 2012;12:2037-45. [Crossref] [PubMed]
- Ahlf Wheatcraft DR, Liu X, Hummon AB. Sample preparation strategies for mass spectrometry imaging of 3D cell culture models. J Vis Exp 2014;(94).
- Li H, Hummon AB. Imaging mass spectrometry of three-dimensional cell culture systems. Anal Chem 2011;83:8794-801. [Crossref] [PubMed]
- Comi TJ, Makurath MA, Philip MC, et al. MALDI MS Guided Liquid Microjunction Extraction for Capillary Electrophoresis-Electrospray Ionization MS Analysis of Single Pancreatic Islet Cells. Anal Chem 2017;89:7765-72. [Crossref] [PubMed]
- Comi TJ, Neumann EK, Do TD, et al. microMS: A Python Platform for Image-Guided Mass Spectrometry Profiling. J Am Soc Mass Spectrom 2017;28:1919-28. [Crossref] [PubMed]
- Wang J, Ren L, Li L, et al. Microfluidics: a new cosset for neurobiology. Lab Chip 2009;9:644-52. [Crossref] [PubMed]
- Majumdar D, Gao Y, Li D, et al. Co-culture of neurons and glia in a novel microfluidic platform. J Neurosci Methods 2011;196:38-44. [Crossref] [PubMed]
- Park J, Koito H, Li J, et al. Microfluidic compartmentalized co-culture platform for CNS axon myelination research. Biomed Microdevices 2009;11:1145-53. [Crossref] [PubMed]
- Kim YT, Karthikeyan K, Chirvi S, et al. Neuro-optical microfluidic platform to study injury and regeneration of single axons. Lab Chip 2009;9:2576-81. [Crossref] [PubMed]
- Southam KA, King AE, Blizzard CA, et al. Microfluidic primary culture model of the lower motor neuron-neuromuscular junction circuit. J Neurosci Methods 2013;218:164-9. [Crossref] [PubMed]
- Ionescu A, Zahavi EE, Gradus T, et al. Compartmental microfluidic system for studying muscle-neuron communication and neuromuscular junction maintenance. Eur J Cell Biol 2016;95:69-88. [Crossref] [PubMed]
- Achyuta AK, Conway AJ, Crouse RB, et al. A modular approach to create a neurovascular unit-on-a-chip. Lab Chip 2013;13:542-53. [Crossref] [PubMed]
- Croushore CA, Sweedler JV. Microfluidic systems for studying neurotransmitters and neurotransmission. Lab Chip 2013;13:1666-76. [Crossref] [PubMed]
- Uzel SG, Platt RJ, Subramanian V, et al. Microfluidic device for the formation of optically excitable, three-dimensional, compartmentalized motor units. Sci Adv 2016;2:e1501429 [Crossref] [PubMed]
- Lee CY, Romanova EV, Sweedler JV. Laminar stream of detergents for subcellular neurite damage in a microfluidic device: a simple tool for the study of neuroregeneration. J Neural Eng 2013;10:036020 [Crossref] [PubMed]
- Tsantoulas C, Farmer C, Machado P, et al. Probing functional properties of nociceptive axons using a microfluidic culture system. PLoS One 2013;8:e80722 [Crossref] [PubMed]
- Brown JA, Pensabene V, Markov DA, et al. Recreating blood-brain barrier physiology and structure on chip: A novel neurovascular microfluidic bioreactor. Biomicrofluidics 2015;9:054124 [Crossref] [PubMed]
- Bear MF, Connors BW, Paradiso MA. Neuroscience: Exploring the Brain. 2nd ed. Philadelphia: Lippincott Williams & Wilkins, 2001.
- Devor M. Unexplained peculiarities of the dorsal root ganglion. Pain 1999;S27-35. [Crossref] [PubMed]
- Hogan QH. Labat lecture: the primary sensory neuron: where it is, what it does, and why it matters. Reg Anesth Pain Med 2010;35:306-11. [Crossref] [PubMed]
- Taylor AM, Blurton-Jones M, Rhee SW, et al. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat Methods 2005;2:599-605. [Crossref] [PubMed]
- Tsutsumi M, Nakatani M, Kumamoto J, et al. In vitro formation of organized structure between keratinocytes and dorsal-root-ganglion cells. Exp Dermatol 2012;21:886-8. [Crossref] [PubMed]
- Klusch A, Ponce L, Gorzelanny C, et al. Coculture model of sensory neurites and keratinocytes to investigate functional interaction: chemical stimulation and atomic force microscope-transmitted mechanical stimulation combined with live-cell imaging. J Invest Dermatol 2013;133:1387-90. [Crossref] [PubMed]
- Bird MM. Establishment of synaptic connections between explants of embryonic neural tissue in culture: experimental ultrastructural studies. Exp Brain Res 1985;57:337-47. [Crossref] [PubMed]
- Vikman KS, Backström E, Kristensson K, et al. A two-compartment in vitro model for studies of modulation of nociceptive transmission. J Neurosci Methods 2001;105:175-84. [Crossref] [PubMed]
- Ohshiro H, Ogawa S, Shinjo K. Visualizing sensory transmission between dorsal root ganglion and dorsal horn neurons in co-culture with calcium imaging. J Neurosci Methods 2007;165:49-54. [Crossref] [PubMed]
- Shipshina MS, Fedulova SA, Veselovskii NS. Induction of Long-Term Depression of Synaptic Transmission in a Co-Culture of DRG and Spinal Dorsal Horn Neurons of Rats. Neurophysiology 2011;43:261-70. [Crossref]
- Phan DT, Bender RHF, Andrejecsk JW, et al. Blood-brain barrier-on-a-chip: Microphysiological systems that capture the complexity of the blood-central nervous system interface. Exp Biol Med (Maywood) 2017;242:1669-78. [Crossref] [PubMed]
- Herland A, van der Meer AD, FitzGerald EA, et al. Distinct Contributions of Astrocytes and Pericytes to Neuroinflammation Identified in a 3D Human Blood-Brain Barrier on a Chip. PLoS One 2016;11:e0150360 [Crossref] [PubMed]
- Fennema E, Rivron N, Rouwkema J, et al. Spheroid culture as a tool for creating 3D complex tissues. Trends Biotechnol 2013;31:108-15. [Crossref] [PubMed]
- Choi YJ, Park J, Lee SH. Size-controllable networked neurospheres as a 3D neuronal tissue model for Alzheimer's disease studies. Biomaterials 2013;34:2938-46. [Crossref] [PubMed]
- Kato-Negishi M, Tsuda Y, Onoe H, et al. A neurospheroid network-stamping method for neural transplantation to the brain. Biomaterials 2010;31:8939-45. [Crossref] [PubMed]
- Park J, Lee BK, Jeong GS, et al. Three-dimensional brain-on-a-chip with an interstitial level of flow and its application as an in vitro model of Alzheimer's disease. Lab Chip 2015;15:141-50. [Crossref] [PubMed]
- Pamies D, Barreras P, Block K, et al. A Human Brain Microphysiological System Derived from Induced Pluripotent Stem Cells to Study Neurological Diseases and Toxicity. ALTEX 2017;34:362-76. [Crossref] [PubMed]
- Lozano R, Stevens L, Thompson BC, et al. 3D printing of layered brain-like structures using peptide modified gellan gum substrates. Biomaterials 2015;67:264-73. [Crossref] [PubMed]
- Kunze A, Giugliano M, Valero A, et al. Micropatterning neural cell cultures in 3D with a multi-layered scaffold. Biomaterials 2011;32:2088-98. [Crossref] [PubMed]
- Turunen S, Haaparanta AM, Aänismaa R, et al. Chemical and topographical patterning of hydrogels for neural cell guidance in vitro. J Tissue Eng Regen Med 2013;7:253-70. [Crossref] [PubMed]
- Huval RM, Miller OH, Curley JL, et al. Microengineered peripheral nerve-on-a-chip for preclinical physiological testing. Lab Chip 2015;15:2221-32. [Crossref] [PubMed]
- Kitagawa Y, Naganuma Y, Yajima Y, et al. Patterned hydrogel microfibers prepared using multilayered microfluidic devices for guiding network formation of neural cells. Biofabrication 2014;6:035011 [Crossref] [PubMed]
- Wang TY, Forsythe JS, Parish CL, et al. Biofunctionalisation of polymeric scaffolds for neural tissue engineering. J Biomater Appl 2012;27:369-90. [Crossref] [PubMed]
- Daud MF, Pawar KC, Claeyssens F, et al. An aligned 3D neuronal-glial co-culture model for peripheral nerve studies. Biomaterials 2012;33:5901-13. [Crossref] [PubMed]
- Rowe L, Almasri M, Lee K, et al. Active 3-D microscaffold system with fluid perfusion for culturing in vitro neuronal networks. Lab Chip 2007;7:475-82. [Crossref] [PubMed]
- Zou Z, Zheng Q, Wu Y, et al. Growth of Rat Dorsal Root Ganglion Neurons on a Novel Self-Assembling Scaffold Containing IKVAV Sequence. Mater Sci Eng 2009;29:2099-103. [Crossref]
- Wei H, Li H, Gao D, et al. Multi-channel microfluidic devices combined with electrospray ionization quadrupole time-of-flight mass spectrometry applied to the monitoring of glutamate release from neuronal cells. Analyst 2010;135:2043-50. [Crossref] [PubMed]
- Mao S, Zhang J, Li H, et al. Strategy for signaling molecule detection by using an integrated microfluidic device coupled with mass spectrometry to study cell-to-cell communication. Anal Chem 2013;85:868-76. [Crossref] [PubMed]
- Dugan CE, Kennedy RT. Measurement of lipolysis products secreted by 3T3-L1 adipocytes using microfluidics. Methods Enzymol 2014;538:195-209. [Crossref] [PubMed]
- Li X, Hu H, Zhao S, et al. Microfluidic Platform with In-Chip Electrophoresis Coupled to Mass Spectrometry for Monitoring Neurochemical Release from Nerve Cells. Anal Chem 2016;88:5338-44. [Crossref] [PubMed]
- Baig NF, Dunham SJ, Morales-Soto N, et al. Multimodal chemical imaging of molecular messengers in emerging Pseudomonas aeruginosa bacterial communities. Analyst 2015;140:6544-52. [Crossref] [PubMed]
- Rizzo DG, Prentice BM, Moore JL, et al. Enhanced Spatially Resolved Proteomics Using On-Tissue Hydrogel-Mediated Protein Digestion. Anal Chem 2017;89:2948-55. [Crossref] [PubMed]
- Chan JH, Timperman AT, Qin D, et al. Microfabricated polymer devices for automated sample delivery of peptides for analysis by electrospray ionization tandem mass spectrometry. Anal Chem 1999;71:4437-44. [Crossref] [PubMed]
- Xu N, Lin Y, Hofstadler SA, et al. A microfabricated dialysis device for sample cleanup in electrospray ionization mass spectrometry. Anal Chem 1998;70:3553-6. [Crossref] [PubMed]
- Meng Z, Qi S, Soper SA, et al. Interfacing a polymer-based micromachined device to a nanoelectrospray ionization Fourier transform ion cyclotron resonance mass spectrometer. Anal Chem 2001;73:1286-91. [Crossref] [PubMed]
- Aerts JT, Louis KR, Crandall SR, et al. Patch Clamp Electrophysiology and capillary electrophoresis-mass spectrometry metabolomics for single cell characterization. Anal Chem 2014;86:3203-8. [Crossref] [PubMed]
- Zhu G, Sun L, Yan X, et al. Bottom-up proteomics of Escherichia coli using dynamic Ph junction preconcentration and capillary zone electrophoresis-electrospray ionization-tandem mass spectrometry. Anal Chem 2014;86:6331-6. [Crossref] [PubMed]
- Choi SB, Lombard-Banek C, Munoz LP, et al. Enhanced peptide detection toward single-neuron proteomics by reversed-phase fractionation capillary electrophoresis mass spectrometry. J Am Soc Mass Spectrom 2018;29:913-22. [Crossref] [PubMed]
Cite this article as: Tillmaand EG, Sweedler JV. Integrating mass spectrometry with microphysiological systems for improved neurochemical studies. Microphysiol Syst 2018;2:3.


