Body-in-a-Cube: a microphysiological system for multi-tissue co-culture with near-physiological amounts of blood surrogate
Introduction
Drug toxicity testing with microphysiological systems (MPS) has the potential to replace animal experiments in the drug development process (1,2). MPS are small microfluidic cell-culture devices that house several tissues as well as a recirculating blood surrogate. Adding drugs to the blood surrogate mimics an intravenous drug administration, and once the drug reaches the co-cultured tissues, it is metabolized via the same pathways that convert it into its metabolites and waste products inside the human body.
Several groups have demonstrated that MPS can replicate known metabolic pathways and produce the expected metabolic products (3-11). Because both efficacy and toxicity depend on drug metabolite concentration profiles, predicting them with MPS requires that metabolite concentrations produced with the systems match those produced in the body.
The main strategies to producing in vivo drug concentration profiles with MPS have been to design the systems so that the ratios of functional units of tissues match those present in vivo (12,13), and to employ tissues of high quality with cellular activities that approach in vivo activities (12). Tissues that approach in vivo cellular activities often consist either of patient-derived primary tissues, or tissues made from stem cells (14-16). Both can be expensive and difficult to obtain in large quantities. MPS that only represent a small fraction of the human body (1/100,000th to 1/50,000th) are best suited for use with such tissues because they require a smaller number of cells to satisfy the functional volume requirement than larger systems. However, in such small systems, the volume of cell culture medium that represents a physiological equivalent of blood is small—60 to 300 µL—and microfluidic systems are difficult to operate with such small amounts of recirculating liquid.
Here we have developed a microphysiological platform that contains chambers for four organs and that can be operated with 80 µL of recirculating cell culture medium. We demonstrate the device with four tissues composed of cells from immortalized cell lines: GI tract epithelial cells (Caco-2), liver cells (HepG2/C3A), bone marrow cells (Meg01), and kidney cells (HK-2). Each tissue was first cultured separately on a 3D scaffold for 24 h, and then the tissues were combined within the device for co-culture. The system supports the co-culture of those tissues for up to 72 h, and recreates acute liver toxicity of acetaminophen and troglitazone. The platform is modular and allows for more tissues to be added in the future. Similar to previous systems (8,17-19), the current design has the added advantage of utilizing gravity to drive fluidic flow, making it inexpensive, reliable, and easy to use.
Methods
MPS design
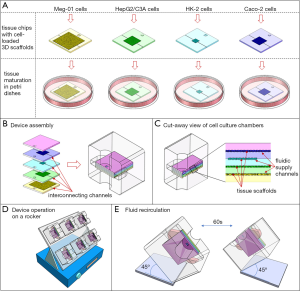
The system consists of a cube-shaped holder that has two medium reservoirs, one on either side, and that can hold four tissue chips (Figure 1). To assemble the MPS, the four tissue chips are first loaded with cells, then stacked on top of each other, and then inserted into the holder. When the chips are stacked on top of each other, the solid back of each chip effectively closes the tissue chamber and fluidic channels of the chip underneath. Each chip has a set of custom-sized microfluidic channels that connect the tissue chamber with the two reservoirs of the holder. When the holder is tilted at a 45° angle, cell culture medium from one reservoir flows through all tissue chips simultaneously, but at individual flow rates. The flow rates are organ-specific and they are determined by the combined hydraulic resistances of the fluidic channels and the cell culture chamber on each chip. The cell culture medium flows into the second reservoir, where it recombines (Figure 1). When the holder is tilted by 45° in the other direction, the recombined cell culture medium now flows back through the tissue chips, exposing all tissues to diluted metabolites. The tilting sequence is repeated every sixty seconds, so that toxic metabolites produced in any of the tissues are redistributed among all tissues.
Tissue volumes
We designed the system with the goal of achieving an overall blood surrogate volume of 80 µL. This volume of blood surrogate corresponds to 1/73000th of the blood volume present in the body of an average person. We designed the cell culture chambers so that they represent 1/73,000th of their in vivo volume. Since blood vessels are represented in our system by microfluidic channels, we subtracted the blood vessel volumes of in vivo organs to obtain the organ volumes that are closer to their actual functional volumes (Vfunctional) (Eq. [1]). The blood vessel content is given by the blood plasma volume (Vplasma), the volume of blood cells (Vblood cells), and the volume of endothelial cells (Vendothelial cells). Data for whole organ volumes and blood vessel content were obtained from Davies et al., Higashi et al., and Price et al. (20-22).
The tissue culture chamber volumes for the cube (Vcube) were obtained by dividing the calculated in vivo functional organ volumes (Vfunctional) by 73000 (scaling factor, SF) (Eq. [2]).
To create 3D tissues within each tissue culture chamber, we seeded cells onto scaffolding that was 200 µm thick and 90% porous. Since the scaffold does not contribute to the function of the tissue, but takes up 10% of the chamber, the chamber volumes were increased by 10%. The final tissue chamber sizes are listed in Table 1.
Table 1
| Organ | Mean in vivo organ volume ± stdev. (20-22) (L) | Functional in vivo organ volume ± stdev. (L) | Cube organ volume ± stdev. (µL) | Square cube chamber area (with chamber depth =200 µm) ± stdev. (mm2) |
|---|---|---|---|---|
| GI tract | 1.23±0.22 | 0.70±0.13 | 9.6±1.7 | 53.3±9.5 |
| Liver | 1.57±0.26 | 0.94±0.16 | 12.8±2.1 | 71.1±11.8 |
| Kidney | 0.32±0.07 | 0.19±0.040 | 2.5±0.53 | 13.9±3.0 |
| Bone marrow | 5.1±0.89 | 2.98±0.52 | 40.8±7.1 | 226.7±29.6 |
| Blood | 5.82±0.73 | 5.82±0.73 | 79.7±10.0 | N/A |
stdev., standard deviation.
Microfluidic channels
Each individual tissue chip contains a set of microfluidic channels that delivers cell culture medium to the tissue chamber. When placed into the holder, the channels line up with the two reservoirs on either side of the chip, forming an interconnected system of tissue chips through which the cell culture medium recirculates (Figure 1).
The channel sizes were chosen to provide a passive hydraulic resistance that limits medium flow through the chip’s tissue chamber to near-physiological values. Calculations of channel sizes followed our earlier method (17), and are outlined below. Flow rates were considered near-physiological when they created fluid residence times within each organ chamber that are comparable to blood residence times in the same volume of tissue in the body (Table 2).
Table 2
| Organ | Qin vivo ± stdev. (20-22) (L/min) | τphys ± stdev. (20-22) (min) | Needed Qcube ± stdev. (µL/min) | Simulated average Qcube ± stdev. (µL/min) |
|---|---|---|---|---|
| GI tract | 0.93±0.16 | 0.75±0.13 | 12.7±2.2 | 11.0±0.1 |
| Liver | 1.30±0.22 | 0.71±0.12 | 18.1±3.1 | 15.6±0.1 |
| Kidneys | 1.20±0.25 | 0.16±0.04 | 16.0±3.3 | 13.8±0.1 |
| Bone marrow | 0.59±0.10 | 5.05±0.87 | 8.1±1.4 | 7.0±0.1 |
stdev., standard deviation.
Blood residence times (τphys) were calculated using the ratio of blood flow through each organ per time interval (Qin vivo) and organ volumes (Vorgan) (Eq. [3]).
We then calculated the needed in vitro flow rate (Qcube) using the functional volumes of the cell culture chambers (Vcube) and the in vivo fluid residence time (τphys) (Eq. [4]).
The needed hydraulic channel resistances that allow us to achieve near-physiological fluid residence times with gravity-driven medium flow are determined via Eqs. [5] and [6]. The channel’s needed hydraulic resistance (R) is calculated using the desired flow rate (Qcube), and the pressure difference (ΔP) between the two medium reservoirs (Eq. [5]).
To create a pressure difference, we tilt the device at an angle so that the liquid levels between the two reservoirs become different. The resulting pressure difference is calculated using the density of the cell culture medium (ρ), the gravitational constant (g), and the resulting height difference between the liquid levels in the two reservoirs (H) (Eq. [6]).
In addition to the hydraulic resistances of the microfluidic delivery channels, we must also consider the hydraulic resistances provided by the channels inside the cell culture chambers. Within each cell culture chamber, the fluidic stream coming from the delivery channel branches out into a set of parallel channels that are 200 µm wide, 200 µm high, and 200 µm apart from each other. The total hydraulic resistance provided by those channels is calculated using Eq. [7]. Here Rn is the hydraulic resistance of each of the n parallel channels that exist in a given cell culture chamber.
The heights and widths of the microfluidic delivery channels can be accurately controlled with microfabrication techniques. The opportunity to adjust those dimensions allows us to create customized hydraulic resistances for each cell culture chamber (Eq. [8]). For channels with rectangular cross-sectional shapes, Eq. [8] relates channel height and width to their hydraulic resistance. We adjust the height and width of each channel to achieve the hydraulic resistance needed to create the desired flow rate (Qcube) in the tissue chamber.
In Eq. [8], η is the kinetic viscosity of the medium, L is a length of the channel, h is the height of the channel, and w is the channel’s width. The calculated and actual organ chamber and channel sizes are listed in Table 3.
Table 3
| Device feature | Organ | Calculated channel/chamber dimensions | Actual channel/chamber dimensions | |||
|---|---|---|---|---|---|---|
| Width (µm) | Height (µm) | Width ± stdev. (µm) | Height ± stdev. (µm) | |||
| Microfluidic channels | bone marrow | 80 | 150 | 75.7±2.2 | 149.1±3.8 | |
| liver | 172 | 176.3±1.7 | ||||
| kidney | 183 | 178.8±2.6 | ||||
| GI tract | 143 | 147.3±2.9 | ||||
| Organ chambers | bone marrow | 15,060 | 200 | 14,859.3±41.3 | 207.9±4.6 | |
| liver | 8,430 | 8,368.3±20.9 | ||||
| kidney | 3,730 | 3,635.7±8.2 | ||||
| GI tract | 7,300 | 7,247.9±18.3 | ||||
stdev., standard deviation.
Fabrication
The cube holder and all tissue chips were designed using 3D drawing software. Each tissue chip measured 20 mm × 20 mm × 1 mm. The tissue culture chambers were designed as 200 µm deep, square cavities with lengths and widths listed in Table 1. The microfluidic channels on the tissue chips were all 200 µm deep, but of varying lengths and widths. The lengths and widths are listed in Table 2. The holder and the tissue chips were all 3D-printed by a commercial vendor using a high-resolution material. The printed tissue chips as well as the chip holder were then coated with a 1 µm thick layer of parylene C. To build the final four-organ system the chips are loaded with cells, stacked against each other, and placed into the cube holder (Figure 1).
Cell culture
HepG2/C3A cells (ATCC) were cultured using Eagl’s Minimum Essential Medium (EMEM) with 50 mL fetal bovine serum. Caco-2 cells (ATCC, HTB-37TM) were cultured using ATCC-formulated Eagle’s Minimum Essential Medium with 100 mL fetal bovine serum. MEG-01 cells (ATCC, CRL-2021TM) were cultured using ATCC-formulated RPMI-1640 medium with 50 mL fetal bovine serum. HK-2 cells (ATCC, CRL-2190TM) were cultured using ATCC-formulated RPMI-1640 medium with 50 mL fetal bovine serum. All cells were maintained at 37 °C, with a volume gas fraction of 5% CO2. Once confluent, the cells were detached from the cell culture flask with trypsin (Trypsin-EDTA, 0.25% volume fraction), and separated from the medium by centrifugation.
Device setup and operation
Tissue culture scaffolds designed for 3D cell culture were obtained from Reprocell USA Inc. (Beltsville, MD). The scaffolds are 200 µm thick, porous, cross-linked polystyrene sheets with an average void size of 42 µm. The scaffolds were loaded with either Caco-2, HepG2/C3A, MEG-01, or HK-2 cells at the following densities: Caco-2: 279×103 cells per scaffold (10% of physiological density), HepG2/C3A: 45×103 cells per scaffold (20% of physiological density), MEG-01: 2,000×103 cells per scaffold (20% of physiological density), HK-2: 12.7×103 cells per scaffold (10% of physiological density). The cell-loaded scaffolds were placed into petri dishes and the cells were maintained for 24 h at 37 °C, with a volume fraction of 5% CO2, using culture medium appropriate for each cell type.
After 24 h, the cell-loaded 3D scaffolds were aseptically transferred into the tissue chambers of the appropriate tissues chip. The chips were then placed on top of each other, so that the back of one tissue chamber closed the opening of the tissue chip below it. The tissue chip stack was then inserted into the holder. The two reservoirs of the cube were filled with 32.5 µL of EMEM (containing a volume fraction of 10% FBS) each, and the device was placed onto a rocker platform that tilted back and forth at an angle of 45°. The platform changed its tilt from 45° to −45° every 60 s continuously for 72 h. Culture medium was collected and replaced with fresh medium after 1, 16, 24, 40, 48, and 64 h. The total amount of blood surrogate (i.e., the amount of cell culture medium that we consider blood surrogate) inside the system consisted of 2×32.5 µL (medium in the reservoirs), plus 15 µL (medium inside the microfluidic channels that deliver medium to the tissue culture chambers).
After 72 h of operation, the cube was disassembled, and the cells on the scaffolds were stained with viability stain (ReadyProbes™ Cell Viability Imaging Kit, Blue/Red, R37610‡).
Urea and albumin concentration measurements
Urea concentrations were measured in the collected cell culture medium using an appropriate assay kit. To conduct the measurement, 5 µL of the collected medium were transferred into the wells of a 96-well plate, and 200 µL of the working reagent was added. The plate was tapped lightly to mix medium and reagents. After 50 min of incubation at room temperature in the dark, the working reagent formed a coloured complex specifically with urea. The absorbance of the coloured complex was measured at 430 nm using a plate reader. The results were obtained from the standard curve and expressed as µg per million cells produced in relation to a 1 h baseline measurement.
Albumin synthesis was evaluated by ELISA (enzyme-linked immunosorbent assay), using a kit and following the manufacturer’s directions. In short, the 96-well plate was coated with goat anti-human albumin antibody and the wells washed with buffer. Diluted samples and standards were added into the coated wells. After incubation, the wells were washed with buffer, horse radish peroxidase-conjugated goat anti-human antibody was added and incubated for 1 h. Following another wash step, 100 µL of enzyme substrate (tetramethylbenzidine) were added and incubated for 15 min. The reaction was stopped by adding 100 µL stop solution. Plates were measured at 450 nm using a plate reader. The results are expressed as µg per million cells produced in relation to a 1 h baseline measurement.
AST measurements
To estimate cell death during co-culture, we measured AST concentrations in the cell culture medium recovered from the devices after 1, 16, 40 and 64 h of co-culture. The AST activity was measured with an assay kit (Sigma MAK055, AST Activity Assay Kit‡). Briefly, standard samples and medium samples were added to the 96 well plate. We then added 100 µL of working solution to each well and mixed both solutions by gently tapping the plate. The plate was then incubated inside a plate reader in the dark, at 37 °C. After 5 min of incubation, the absorbance at 450 nm was measured, and the measurement was repeated every 5 minutes until absorbance of the most active sample exceeded the standard curve’s largest absorbance. We then selected the initial absorbance value inside the linear range and calculated the AST concentrations using the protocol given in the kit. The results are expressed as µg per million cells produced in relation to a 1 h baseline measurement.
Cell number measurement
The cell number in each tissue culture scaffold before culture on the chip was measured by alamarBlue™ HS Cell Viability Reagent (ThermoFisher Scientific, A50101‡). Briefly, the standard curve for each cell line was obtained by seeding predetermined numbers of cells (counted by Trypan Blue using Countess™ II Automated Cell Counter, AMQAX1000, ThermoFisher Scientific‡) into 6 well plates and measuring the fluorescence of incubated culture medium (ex/em is 560/590). The cell numbers on the seeding scaffold was measured following a similar procedure by incubation with alamar blue reagent and measurement of the incubated culture medium. After co-culture experiments, cell viability was determined by cell viability stain (ReadyProbes™ Cell Viability Imaging Kit, Blue/Red, R37610‡)
Flow rate measurement
To determine the volume flow rate, we added 5 µL EBM-2 with growth factor to the bottom reservoir, then 65 µL of EBM-2 with growth factor to the top reservoir. We then let the medium flow through the device for thirty seconds. The culture medium in the bottom reservoir was collected and weighed. The volume change of culture medium in the bottom reservoir and volume flow rate was calculated. Then the flow rate was adjusted to account for the viscosity difference at room temperature and 37 °C.
Computational simulation of flow dynamics
We simulated the fluidic flow inside the body cube using a method we described earlier. In short, 3D software models of the tissue chambers were imported into COMSOL 5.5‡. Stationary total flow rates in the device under 6 different liquid level differences were simulated in COMSOL. The correlation of the liquid level differences and total flow rate was determined by polynomial regression curve fitting. Then one partial differential equation (PDE) for the total flow rate in the device was built based on the liquid level difference versus flow rate fitting curve, and another PDE for the angular position of the device was built based on the motion of the rocking platform. Those PDEs were solved with MATLAB PDE solver ode45 with absolute tolerance 10−10 and a relative tolerance 10−7 in MATLAB R2016b‡. Since the ratios of flow rate in each organ chamber were equal to the ratios of the reciprocal of their hydraulic resistances, the flow rate in each chamber could be calculated.
Results
System design
To accommodate four tissues with 80 µL of recirculating cell culture medium, we designed a cube-shaped holder that held a total of four tissue chips (Figure 1). Stacking the chips allowed us to shorten the interconnects and with that the amount of liquid needed to operate the device. Medium flow across the tissues was achieved via channels that were etched into the tissue culture chamber lids, i.e., the back of each adjacent tissue chip. The channels were 200 µm wide on a 400 µm pitch.
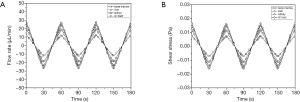
The amount of recirculating cell culture medium inside the body cube consisted of three fractions. The combined amount of cell culture medium in the two reservoirs was 65 µL at any given time, while the amount of cell culture medium inside the network of fluidic channels was 15 µL. The system also contained a small amount of cell culture medium that resided in each tissue culture chamber and filled the space not occupied by either cells or scaffold. The total flow rate measured was 3.36±0.40 µL/s. The flow in each tissue culture chamber was simulated computationally and is shown in Figure 2. The flow rate periodically increases and then reverses direction as the device is rocked back and forth. The average flow rates were 11.0±0.1 µL/min (GI tract), 15.6±0.1 µL/min (liver), 13.8±0.1 µL/min (kidney), and 7.0±0.1 µL/min (bone marrow).
Cell viability and function
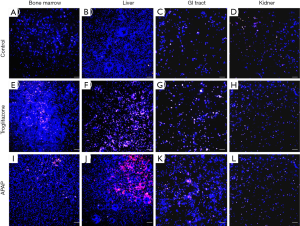
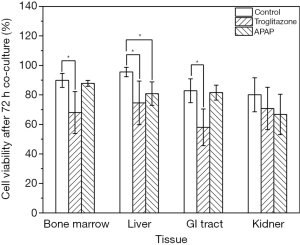
We recovered all tissue scaffolds from the cube after 72 h of co-culture, stained the cells with viability dyes, and imaged the cells via fluorescence microscopy (Figure 3). The images show that the scaffolds recovered from all four tissue chambers are populated with live cells as well as a smaller fraction of dead cells. Bone marrow and liver cells were most amenable to 72 h of co-culture in the low-liquid environment. Image analysis showed that 95.5%±3.2% of liver cells, and 89.8%±4.7% of bone marrow cells were still viable after the recovery (Figure 4). The numbers of live cells after 72 h of operation were lower in the GI tract tissue (82.8%±8.1%) and the kidney tissue (80.1%±11.5%) (Figure 4).
To measure the production and secretion of albumin and urea into the cell culture medium, we used a portion of the cell culture medium recovered from the devices every day. On the first day of culture, the medium albumin content was 0.74±0.11 µg per day per million cells, with lower concentrations measured on days two (0.55±0.13 µg per day per million cells) and three (0.31±0.04 µg per day per million cells). Similarly, the urea content in the cell culture medium was 72.1±6.0 µg per day per million cells on the first day, and lower on days two (64.6±9.8 µg per day per million cells) and three (53.4±10.5 µg per day per million cells) of co-culture.
Toxicity measurements
To determine whether the body cube can detect acute cellular toxicity, we challenged the device with two toxins that are known to cause damage to liver HepG2/C3A cells when exposed at high concentrations, acetaminophen (23), and troglitazone (24,25). Similar to the baseline experiments described above, we recovered cell cultures after 72 h of co-culture from the device. Fluorescence images confirm that both acetaminophen and troglitazone cause significant liver damage when compared to control conditions without drug (Figures 3,4). This result is confirmed by the differences in AST released from cells cultures treated with acetaminophen and troglitazone. Compared to control conditions without drug, the amount of AST in the cell culture medium is significantly higher, indicating significant liver cell death. In addition, urea production, but not albumin production was decreased when drugs were added to the cube (Figure 5).

Discussion
System design
Our goal was to design a microphysiological system that can be operated with small amounts of cell culture medium, so that the volume of blood surrogate in the system is close to physiological values. We calculated on-chip organ volumes and on-chip blood surrogate content using data for a 70 kg male human (20-22), and when necessary, we normalized data from the literature to a 70 kg human by scaling the values assuming a directly proportional relationship. Because organs contain varying amounts of vasculature, and the in vitro tissues we constructed here do not (the blood vessels are moved to the outside of the tissue and are mimicked by the fluidic channels that supply cell culture medium to the tissues), we removed the volumes of vascular endothelial cells and blood volumes from the overall reported organ volume to obtain a volume that is closer to its functional volume. The amount of blood surrogate volume (80 µL) was calculated using the same scaling factor (73,000) as for the organ chambers. Our method to calculate functional organ volumes is only a first approach to creating MPS, and a more detailed analysis of functional organ volumes according to Wikswo et al. (13) could provide an even more accurate system in the future.
To operate the MPS with only physiological amounts of cell culture medium (80 µL), we designed the system in a cube format where the tissue chips are stacked on top of each other, and fluidic connections among organ chambers are short. In addition, using gravity to drive fluidic flow allowed us to eliminate tubing and connections typically used with peristaltic pumps, and with that to decrease the amount of liquid needed to operate the system.
The device we developed here is modular, meaning that all tissue chips can be handled separate from each other during the time of cell seeding. Tissue maturation can take place in separate dishes with cell culture medium customized for each cell type. Each chip also contains channels with tissue-specific dimensions that provide the connection to the main reservoirs. This feature allows us to quickly adjust the overall system by switching organ chips, and adding new ones when needed.
Liquid content
Physiological amounts of blood surrogate (cell culture medium) in an MPS are relatively small amounts of liquid, making it difficult to recirculate such a surrogate in conventional 2D microfluidic systems. In the MPS presented here, all organ compartments were scaled by a factor of 73,000, and the corresponding physiological amount of blood surrogate is 80 µL of cell culture medium. Our goal with this work was to demonstrate an MPS design that can recirculate such small amounts of cell culture medium.
To operate the MPS with 80 µL of cell culture medium, we designed it as a cube. In the cube, interconnecting channels needed to recirculate the liquid among all cell culture chambers are short, allowing us to stay within the 80 µL limit. All conventional MPS have a 2D layout, where organ chambers are arranged next to each other on a 2D plane. The channels that interconnect organ chambers with each other and with medium reservoirs are long and require additional liquid volumes that push the amount of blood surrogate within the system beyond what would be physiological.
An alternative strategy to achieve physiological amounts of blood surrogate in an MPS is to scale the organ chambers in the device less aggressively using a smaller scaling factor. The overall volumes of tissues and blood surrogate would increase, making it easier to operate and handle the MPS. However, in order to retain physiological tissue cell densities, that strategy would require the use of larger numbers of cells to construct each tissue. When using patient-derived primary cells, using larger numbers of cells may become prohibitively expensive. Choosing scaling factors between 60,000 and 100,000 are likely the most useful, because the resulting cell culture chambers can be filled with in vitro tissue constructs containing less than a million cells. At the same time, the blood surrogate volume would still be within a range that can be recirculated (60 to 100 µL).
In general, another approach to achieving physiological liquid volumes in MPS is to eliminate tissue chambers for tissues that are known to not interact with the drug to be tested. Such tissues would neither absorb, redistribute, or convert the drug, or be otherwise affected by it. When this is the case, the chamber for that organ can be eliminated along with the accompanying fluidic channels, decreasing the amount of liquid needed to operate the MPS.
Despite containing physiological amounts of blood surrogate, the overall liquid-to-cell ratio in our device is still not fully physiological. This is in large part due to the tissue’s cell densities that range between 10% to 20% of in vivo values (Table 4). Similar to the tissues in the human body, the spaces in each organ chamber that are not occupied by either cells or scaffold, are also filled with cell culture medium. The amount of that interstitial liquid, i.e., the liquid between cells, can also significantly contribute to the dilution of drug metabolites. When using in vitro cell cultures, attention must be paid to the density of cells achieved within the tissue construct. In vitro tissues tend to be much less densely populated with cells than in vivo tissues. That means the amount of interstitial liquid is higher than it would be in the body. While the cell density within the tissue construct we used was still far from physiological values (10% to 20%), future developments of 3D scaffolds that allow for higher cell densities will help eliminate non-physiological amounts of interstitial liquid.
Table 4
| Physiological and seeded cell numbers | Liver | Bone marrow | GI tract | Kidney |
|---|---|---|---|---|
| Physiological cell number per tissue chamber | 2,790×103 | 10,000×103 | 222×103 | 127×103 |
| Number of seeded cells (percentage of physiological cell number) | 279×103 (10%) | 2,000×103 (20%) | 44.4×103 (20%) | 12.7×103 (10%) |
Cell viability and metabolism
Our device contained 80 µL of liquid, and supported four tissues for three days. Fluorescent images of scaffolds with cells recovered from the cube after 72 h of operation were evaluated and confirm that the majority of cells were viable in all cell culture chambers. Some cells (5% to 20%, depending on the tissues) were no longer viable, similar to what Sung et al. have measured with their device without renewal of cell culture medium (26).
Liver cells also produced albumin and urea in quantities similar to those observed in other in vitro devices (27), with values showing a downward trend at the 72 h timepoint. A 24 to 72 h timeframe is suitable for detecting acute drug toxicity, but chronic effects require longer co-culture times to manifest.
To achieve co-culture times that are longer than three days, additional strategies to maintain the cultures with low levels of liquid need to be developed. First, evaporation of liquid must be limited. While using our body-cube we noticed that a significant amount of cell culture medium (about 10%) evaporated from the system per day. This decrease in liquid level is a consequence of operating the system with gravity, which means the system is not fully closed and cell culture medium is exposed to the incubator environment. That exposure enables necessary gas exchange, but also permits evaporation. When a significant amount of cell culture medium evaporates, concentrations of cellular waste products increase, which could affect cell viability.
In order to limit the effects of evaporation, we replaced the entire cell culture medium every 8 to 16 hours, a practice that also allowed us to remove waste products. However, replacing the cell culture medium every day influences the concentration profiles of any added drugs, as well as those of drug metabolites. In the future, instead of replacing the medium, it would be preferable to include a mechanism that allows for waste removal in another way. A functioning kidney tissue would serve that purpose and likely also increase the time cells are viable. We believe that without such a mechanism the usefulness of microphysiological devices will be limited to evaluating the effects of only acute 24 h drug exposures.
An additional consideration that could allow the cube to achieve longer cell culture times is to improve the composition of the cell culture medium. Here, we used a composition that consisted of equal parts of cell culture medium optimized for each of the four cell types. A custom formulation designed specifically to support all four tissues at the same time could help limit the detrimental effects of limited availability of specific nutrients or growth factors.
Toxicity measurements
To demonstrate that the body cube is capable of measuring drug induced liver injury, we challenged the device with two drugs that are known to cause liver cell toxicity in both primary cells and HepG2 cells (acetaminophen) (23) and in HepG2 cells only (troglitazone) (24,25). Both drugs caused the expected cell death, and in part due to the decrease in cell number, we also observed an accompanying decrease in metabolic activity of the tissues. Similarly, cytosolic enzymes are released in significantly larger amounts from cell cultures challenged with both drugs than those that were not exposed to drugs after 40 h of exposure. After 48 h the amount of cytosolic enzyme concentration in the medium is similar for both drug-exposed cultures and control cultures. The downward trend in drug-exposed cultures after 40 h is likely due to the fact that a large number of the most sensitive cells were already damaged earlier. On the other hand, the upward trend in control cultures is likely due to the limit in culture time in the cube. The result highlights the need to lengthen the time of tissue viability in the cube. In addition, to detect liver cell toxicity of troglitazone to primary liver cells, future experiments must include 3D tissues of primary liver cells in combination with other non-parenchymal liver cells as demonstrated by Kostadinova et al. (28).
Limitations of the body cube
The main shortcoming of the body cube is that while stacking tissue chips on top of each other shortens fluidic connections, it also means that each individual chip is no longer accessible via optical microscopy. Optical microscopy, on the other hand, is one of the most common methods used to evaluate tissue health during and after microphysiological device operation. To obtain images of tissues and other visual data that communicate such information, integrated optical elements and sensors will be needed in the future. Similarly, integrated flow sensors are needed to verify fluidic flow within each tissue chamber and the overall microfluidic connections.
A second limitation with the current design is that the flow created via gravity is bidirectional, causing shear in two directions. At low shear values, cells of many tissues, including liver tissue, are not affected adversely (27). However, even low shear can create pro-inflammatory conditions in endothelial cells when the flow is bidirectional (29). Endothelial cells are key to simulating the uptake of a drug. Without the endothelium, microphysiological devices can still be used to conduct proof-of-concept trials, but they will not deliver entirely accurate readings of drug toxicity. Future devices should be designed with valves that create unidirectional flow, similar to those published earlier (17,30). In addition, those integrated valves must also be designed with small operating volumes so that near-physiological levels of blood surrogate are maintained.
Although the total physiological flow rate for a system scaled by 1/73,000th is 0.92 µL/s, the flow rate in our system was about three times as high. This is likely due to additional flow that occurred through the spaces in the cell culture chambers that were not occupied by tissue scaffold or actual tissue. In the future, the culture of the endothelial barrier tissue between tissues and fluidic channels could provide an additional barrier to prevent this type of flow.
Conclusions
The body cube is a multi-organ microphysiological device that can be operated with small, near-physiological amounts of blood surrogate (cell culture medium). Cells of four tissues, cultured in the cube for three days, were viable and functional, indicating that the cube can be used to test for the toxicity drugs and its metabolites during an acute 24 to 72 h drug exposure. The developed cube is modular, and was operated with gravity-induced flow, making it easy to use, and attractive for large-scale drug toxicity studies. Other groups have focussed on increasing the number of tissues and using more physiologically relevant types of cells in microphysiological devices. Because it is a key issue in delivering microphysiological devices that predict drug toxicity accurately, we have here focused on strategies to limit excess liquid.
Acknowledgments
HU is a Research Fellow of the Japan Society for the Promotion of Science and was funded by the society. Nanofabrication was done at the NIST Nanofabrication Facility at the Center for Nanoscale Science and Technology. LC acknowledges support under the Cooperative Research Agreement between the University of Maryland and the National Institute of Standards and Technology Physical Measurement Laboratory, Award 70NANB14H209, through the University of Maryland.
Funding: The article was supported in part by 999-NIST, Official duty at NIST DOC.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/mps-19-8). The University of Maryland has filed a provisional patent for the described technology with MBE and LC as inventors. HU and YY have no conflicts of interest to declare.
Disclaimer: Certain commercial materials are identified in this paper to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement by the National Institute of Standards and Technology, nor is it intended to imply that the materials or equipment identified are necessarily the best available for the purpose.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
‡any mention of commercial products within this work is for information only. It does not imply recommendation or endorsement by NIST.
References
- Baker M. A living system on a chip. Nature 2011;471:661-5. [Crossref] [PubMed]
- Esch MB, Mahler GJ. Chapter 11 - Body-on-a-chip systems: Design, fabrication, and applications. In: Borenstein JT, Tandon V, Tao SL et al. editors. Microfluidic Cell Culture Systems (Second Edition). Cambridge: Elsevier, 2019:323-50.
- Viravaidya K, Shuler ML. Incorporation of 3T3-L1 Cells To Mimic Bioaccumulation in a Microscale Cell Culture Analog Device for Toxicity Studies. Biotechnol Prog 2004;20:590-7. [Crossref] [PubMed]
- Tatosian DA, Shuler ML. A novel system for evaluation of drug mixtures for potential efficacy in treating multidrug resistant cancers. Biotechnol Bioeng 2009;103:187-98. [Crossref] [PubMed]
- Mahler GJ, Esch MB, Glahn RP, et al. Characterization of a gastrointestinal tract microscale cell culture analog used to predict drug toxicity. Biotechnol Bioeng 2009;104:193-205. [Crossref] [PubMed]
- Esch MB, Smith AST, Prot JM, et al. How multi-organ microdevices can help foster drug development. Adv Drug Deliv Rev 2014;69-70:158-69. [Crossref] [PubMed]
- Prot JM, Bunescu A, Elena-Herrmann B, et al. Predictive toxicology using systemic biology and liver microfluidic “on chip” approaches: Application to acetaminophen injury. Toxicol Appl Pharmacol 2012;259:270-80. [Crossref] [PubMed]
- Oleaga C, Bernabini C, Smith AST, et al. Multi-Organ toxicity demonstration in a functional human in vitro system composed of four organs. Sci Rep 2016;6:20030. [Crossref] [PubMed]
- Oleaga C, Riu A, Rothemund S, et al. Investigation of the effect of hepatic metabolism on off-target cardiotoxicity in a multi-organ human-on-a-chip system. Biomaterials 2018;182:176-90. [Crossref] [PubMed]
- Theobald J, Ghanem A, Wallisch P, et al. Liver-Kidney-on-Chip To Study Toxicity of Drug Metabolites. ACS Biomaterials Science & Engineering 2018;4:78-89. [Crossref]
- Bovard D, Sandoz A, Luettich K, et al. A lung/liver-on-a-chip platform for acute and chronic toxicity studies. Lab Chip 2018;18:3814-29. [Crossref] [PubMed]
- Edington CD, Chen WLK, Geishecker E, et al. Interconnected Microphysiological Systems for Quantitative Biology and Pharmacology Studies. Sci Rep 2018;8:4530. [Crossref] [PubMed]
- Wikswo JP, Curtis EL, Eagleton ZE, et al. Scaling and systems biology for integrating multiple organs-on-a-chip. Lab Chip 2013;13:3496-511. [Crossref] [PubMed]
- Zeilinger K, Freyer N, Damm G, et al. Cell sources for in vitro human liver cell culture models. Exp Biol Med (Maywood) 2016;241:1684-98. [Crossref] [PubMed]
- Pan C, Kumar C, Bohl S, et al. Comparative Proteomic Phenotyping of Cell Lines and Primary Cells to Assess Preservation of Cell Type-specific Functions. Mol Cell Proteomics 2009;8:443-50. [Crossref] [PubMed]
- Wilkening S, Stahl F, Bader A. Comparison of primary human hepatocytes and hepatoma cell line HEPG2 with regard to their biotransformation properties. Drug Metab Dispos 2003;31:1035-42. [Crossref] [PubMed]
- Esch MB, Ueno H, Applegate DR, et al. Modular, pumpless body-on-a-chip platform for the co-culture of GI tract epithelium and 3D primary liver tissue. Lab Chip 2016;16:2719-29. [Crossref] [PubMed]
- Chen HJ, Miller P, Shuler ML. A pumpless body-on-a-chip model using a primary culture of human intestinal cells and a 3D culture of liver cells. Lab Chip 2018;18:2036-46. [Crossref] [PubMed]
- Lee H, Kim DS, Ha SK, et al. A pumpless multi-organ-on-a-chip (MOC) combined with a pharmacokinetic–pharmacodynamic (PK–PD) model. Biotechnol Bioeng 2017;114:432-43. [Crossref] [PubMed]
- Davies B, Morris T. Physiological Parameters in Laboratory Animals and Humans. Pharm Res 1993;10:1093-5. [Crossref] [PubMed]
- Higashi Y, Noma K, Yoshizumi M, et al. Endothelial Function and Oxidative Stress in Cardiovascular Diseases. Circ J 2009;73:411-8. [Crossref] [PubMed]
- Price PS, Conolly RB, Chaisson CF, et al. Modeling Interindividual Variation in Physiological Factors Used in PBPK Models of Humans AU - Price, Paul S. Crit Rev Toxicol 2003;33:469-503. [Crossref] [PubMed]
- Hinson JA, Roberts DW, James LP. Mechanisms of Acetaminophen-Induced Liver Necrosis. In: Uetrecht J, editor. Adverse Drug Reactions. Berlin, Heidelberg: Springer Berlin Heidelberg, 2010:369-405.
- Tirmenstein MA, Hu CX, Gales TL, et al. Effects of Troglitazone on HepG2 Viability and Mitochondrial Function. Toxicol Sci 2002;69:131-8. [Crossref] [PubMed]
- Masubuchi Y, Kano S, Horie T. Mitochondrial permeability transition as a potential determinant of hepatotoxicity of antidiabetic thiazolidinediones. Toxicology 2006;222:233-9. [Crossref] [PubMed]
- Sung JH, Shuler ML. A micro cell culture analog (µCCA) with 3-D hydrogel culture of multiple cell lines to assess metabolism-dependent cytotoxicity of anti-cancer drugs. Lab Chip 2009;9:1385-94. [Crossref] [PubMed]
- Esch MB, Prot JM, Wang YI, et al. Multi-cellular 3D human primary liver cell culture elevates metabolic activity under fluidic flow. Lab Chip 2015;15:2269-77. [Crossref] [PubMed]
- Kostadinova R, Boess F, Applegate D, et al. A long-term three dimensional liver co-culture system for improved prediction of clinically relevant drug-induced hepatotoxicity. Toxicol Appl Pharmacol 2013;268:1-16. [Crossref] [PubMed]
- Yang Y, Fathi P, Holland G, et al. Pumpless microfluidic devices for generating healthy and diseased endothelia. Lab Chip 2019;19:3212-9. [Crossref] [PubMed]
- Wang YI, Shuler ML. UniChip enables long-term recirculating unidirectional perfusion with gravity-driven flow for microphysiological systems. Lab Chip 2018;18:2563-74. [Crossref] [PubMed]
Cite this article as: Chen L, Yang Y, Ueno H, Esch MB. Body-in-a-Cube: a microphysiological system for multi-tissue co-culture with near-physiological amounts of blood surrogate. Microphysiol Syst 2020;4:1.