
Recent advancements of human iPSC derived cardiomyocytes in drug screening and tissue regeneration
Introduction
The heart, one of the most important organs, provides adequate blood flow to other organs and tissues with coordinated contractions. However, the inability of cardiac tissue to regenerate and the inability of cardiomyocytes (CMs) to amplify in vitro makes the cardiac muscle unable to self-heal after myocardial infarction. The only way to replace any infarct myocardium, which can result from hypertension or heart attack, is by heart transplant. Nonetheless, there are major limitations associated with heart transplantation, including a shortage of organ donors, the need for immunosuppressive medications and problems regarding medical ethics. For this reason, the differentiation of human-induced pluripotent stem cells (hiPSCs) into CMs (hiPSC-CMs) is regarded as one of the most promising alternative treatments for the regeneration of damaged cardiac tissues after myocardial infarction.
HiPSC differentiation was first developed by Takahashi et al., who used a combination of four transcription factors (Oct4, Sox2, c-Myc, and Klf4) to differentiate somatic cells into embryonic stem (ES)-like cells (1). Another group of factors (Oct4, Sox2, Nanog, and Lin28) was used for iPSC lines through reprogramming human somatic cells. These kinds of stem cells also had very similar stem cell markers and telomerase activity with embryonic stem cells (ESCs) (2). On the other hand, Moretti et al. infected dermal fibroblasts using retroviral vectors with encoded transcription factors to generate patient-specific pluripotent stem cells, leading to the possibility of establishing reliable human cardiac disease models (3).
Ever since they were first developed, hiPSCs have had promising prospects in medical screening. Currently, animal models such as mice or rats are the most common models in drug screening. However, interspecies differences can lead to many inconsistencies in basic research and clinical trials. Because many diseases are affected by several genetic factors, such as long QT syndrome which is caused by hERG (human Ether-a-go-go Related Gene), it is difficult to establish human-specific disease models using rodent model organisms. The reason why hiPSCs are opening up a new era of translational medicine is because they are not only capable of reconstructing the specific pluripotent cell lines of individual patients, but are also capable of reproducing disease phenotypes. Nonetheless, hiPSC derived CMs are still in the early stages of development. Many pieces of research have highlighted the potential of utilizing iPSC-CMs for cardiac repair, since iPSC-CMs have the potential to become an ideal tool for the screening of therapeutic agents and cardiac cytotoxicity, and their functions can be detected by recording spontaneous and electrical field-stimulated contractions (4,5). On the other hand, disease-specific hiPSC-CMs can be used to test cardiotoxic drug reactions by measuring their action potential duration through a variety of methods, such as early after depolarization (6).
HiPSC-CMs have also had exciting potential applications in personalized medicine. Personalized medicine is a customized model that designs the most appropriate treatments or products based on personal genomic information and relevant internal environment information, resulting in treatments with maximum effects and minimal side effects (7). However, in clinical practice, individual differences among patients greatly impede the drug discovery process and reduce therapeutic effects (8,9). One of the reasons this may happen could be the minor genetic differences between each patient that may contribute to personalized cardiac phenotypes and pathological mechanisms. Therefore, hiPSC-CMs have emerged as an example of a new therapy for serious heart diseases because of their unique advantages, such as their ability to act as a patient-specific cell source, or their ability to create in vitro models of human pathophysiology (10-12). iPSCs are a promising source for constructing personalized heart models for patient-specific drug screening and disease mechanism research, since iPSCs are derived in a patient-specific manner. Meanwhile, the increasingly sophisticated approaches for generating iPSC lines from human cells and the improvements in high through-put physiological assays have laid the foundation for the development of patient-specific disease modeling platforms for pre-clinical trials (13). Although hiPSCs have advantages over conventional cell lines and animal models, they do not fully reflect the drug’s effect on individuals suffering from disease development and environmental factors. With the rapid development of tissue engineering, the organs-on-chip technologies are expected to be widely used in disease modeling. Despite the enormous potential hiPSCs have for clinical applications, the efficiency and purity of the differentiation have remained as decade-long challenges for scientists.
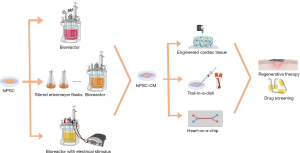
Recently, several critical reviews have been published about the characterization of differentiated iPSC-CMs, as well as their applications in regenerative therapy, disease modeling and drug screening. Karakikes et al. focused on the molecular, cellular and functional phenotypes of iPSC-CMs, which could decide the clinical application potential of iPSC-CMs (14). Masumoto et al. concluded their research about cell sheet technologies using temperature-responsive culture surfaces and iPSC-CMs, which presented high differentiation efficiency, for cardiovascular regeneration (15). Also, the therapeutic effects of human pluripotent stem cell (hPSC) derived CMs on myocardial injuries and bioengineering approaches to enhance therapeutic effects were summarized by Park et al. (16). Both Smith et al. and del Álamo et al. proposed iPSC-CMs as a promising platform for disease modeling and drug screening, especially high throughput screening (17,18). Unlike recently published reviews, the current review not only introduces various methods to improve the differentiation efficiency of iPSC-CMs, but also discusses various subtypes of obtained iPSC-CMs and the differences in electrophysiological characteristics between iPSC-CMs and matured native CMs. Furthermore, large-scale differentiation and expansion methods are discussed for achieving biomanufacturing strategies. These are combinatorial approaches with biomaterials and advanced microfabrication methods to engineer iPSC-CMs for creating biomimetic 3D cardiac tissue constructs. Particularly, recent advancements in the application of engineered hiPSC-CMs in cell-free regenerative therapy and heat-on-a-chip for drug screening are also presented. Biomimetic strategies for iPSC-CM laboratory research and personalized clinical applications are systematically introduced here.
Differentiation and Electrophysiological characteristics of hiPSC-CMs
The differentiation of CMs from hiPSCs is a committed step in creating artificial cardiac muscle cells and tissues. In this section, various methods to induce the differentiation of hiPSC-CMs, as well as engineering approaches to purify and amplify the hiPSC-CMs, are described. Conceptually, the differentiation of iPSCs is no different than that of embryonic stem cells (ESCs). The differentiation of both iPSCs and ESCs follow the same general procedures (19,20). Therefore, we include some ESC differentiation methods which work for iPSCs as well. The differentiation of hiPSC-CMs makes it possible to obtain patient-specific CMs from adult somatic cells, but hiPSC-CMs are still not identical to original mature CMs. We also summarize the differences between hiPSC-CMs and original mature CMs in terms of electrophysiological characteristics and various subtypes in this section.
Method to obtain a large quantity of CMs from iPSCs
Following the methods to differentiate human embryonic stem cells (hESCs), early attempts to differentiate hiPSCs used the spontaneous, embryonic body (EB)-based, CM differentiation system. In these studies, hiPSCs were transferred to suspension cultures, forming three-dimensional (3D) EBs, and then plated on culture dishes coated by gelatin to differentiate. The generated hiPSC-CMs were similar to early stage human CMs in terms of structural and functional properties (21,22). However, the above-mentioned spontaneous EB-based differentiation led to various types of somatic cells being generated, indicating a low conversion ratio and a low purity of hiPSC-CMs. Only 1% to 10% of hPSCs yielded CMs with a conventional EB-based differentiation system.
Inspired by the differentiation that happens in vivo, researchers have utilized a variety of factors that affect in vivo differentiation to regulate the differentiation of hPSC-CMs, resulting in up to 70% of hPSCs yielding CMs (23). Growth factors including the wingless/INT proteins (WNTs), the transforming growth factor-β superfamily (TGF-β), and the fibroblast growth factors (FGFs) have been confirmed to induce specialized differentiation of CMs from hESCs (24-27). By optimizing the concentrations of serum, polyvinyl alcohol, bone morphogenetic protein-4 (BMP-4) and FGF2, together with insulin in a chemically defined medium, contracting human EBs (hEBs) composed of high proportions of cardiac troponin I positive cells were obtained (28). Due to the similarity of hiPSCs and hESCs, most of the above growth factors work for the differentiation of hiPSCs as well (29). The co-culture of hPSCs with END-2 cells or Visceral Endoderm-Like Cells also enhances the differentiation efficiency of hPSC-CMs, which may result from paracrine signaling (30,31).
For improving the differentiation efficiency of hPSC-CMs using growth factors, cells grow as two-dimensional (2D) monolayers that growth factors can uniformly distribute. Later, this monolayer culture method combined with growth factors improved the maturation of CMs, allowing for larger-scale differentiations compared to EB-based CM differentiation approaches (32). By combining a variety of components with monolayers, researchers have increased the purity of CMs dramatically. The combination of activin A and BMP-4 in a serum-free monolayer medium has generated a total of over 50% of contracting CMs (33). Based on this, some universal cardiac differentiation systems were developed. The application of dynamic extracellular matrices in monolayer cultures promoted the epithelial-mesenchymal transformation of hPSCs and produced CMs of up to 98% purity (34). Temporal modulation of classic WNT signaling regulators could produce 0.8–1.3 million functional CMs per cm2 of high purity (80–98%) (35). The combined use of KY02111 and WNT signaling regulators could create robust cardiac differentiation in the absence of any types of cytokines and hormones, such as the previously mentioned BMP-4, Activin A or insulin (36). Here, we summarized a systematic comparison of some mainstream differentiation protocols (Table 1).
Table 1
| Method | Differentiation protocols | Resulting hiPSC-CMs | Ref | ||||||
|---|---|---|---|---|---|---|---|---|---|
| DIVs | Induction factors | Efficiency | Purity | Electrophysical functions | Beat rate | Major expressed markers | |||
| Embryoid Bodies | 8–9 days | Suspension culturing | – | About 10% | Exhibiting the nodal, atrial, and ventricular phenotypes | 41–47 bpm | NKX2-5, TNNT2, MYH6, ACTN2, MYL7, MYL2, HPPA, PLN | (21) | |
| 16 days | Suspension culturing+0.1% gelatin-coated dish | – | – | Stable pacemaker activity and synchronized action potential propagation | 21 bpm | sarcomeric α-actinin, cardiac troponin-I, Cx43, cardiac-specific transcriptional factors (GATA4, MEF-2c, NKX2.5), Sarcomeric protein (MYH06, MYH-7MLC-2v, Ctnni), ion channels (CACNA1C, CACNA1D, KCNH2, KCNQ1) | (22) | ||
| Embryoid molecules | 26 days | WNT signaling molecules | – | 90% | – | – | α-actinin, cTNT, Nkx2.5 | (23) | |
| Monolayer | 9 days | BMP-4 + FGF2 + polyvinyl alcohol + serum + insulin | 91.2±1.9% | 64–89% | Reproducible action potential duration and conduction velocities | – | MESP1, NKX2-5, ISL1, TNNT2, MYH6, ACTN2, TNNI3, CX43 | (28) | |
| Tissue engineering | 25–31 days | 3D engineered tissue + WNT signaling | – | – | Promotes INa density and action potential upstroke velocity | – | INa, α-actinin, Nav1.5 | (37) | |
| chemical defined | 11–13 days | Basal medium RPMI 1640, L-ascorbic acid 2-phosphate and rice-derived recombinant human albumin | – | 80–95% | An atrial-like action potential with a low average maximum diastolic potential (MDP) of −55.2±2.0 mV | – | TNNT2, α-actinin | (32) | |
While the above methods are effective, they involve the use of various growth factors that are expensive and require long periods of time for concentration optimization. Many researchers have been dedicated to the simplification of differentiation systems for hiPSC-CMs without the use of growth factors. The first fully chemical-defined platform was developed to provide highly reproducible differentiation, which was responsible for furthering the understanding of the needed macromolecules. This simplified platform consisted of only three components: RPMI 1640, L-ascorbic acid 2- phosphate and rice-derived recombinant human albumin (32).
The application of hiPSCs for clinical therapies requires large-scale expansion (Figure 1) and a controlled differentiation process. However, conventional production methods, such as monolayer culture surfaces, offer limited surface and require repeated sub-culturing, making it difficult to adequately supply a large number of cells. Cell reprogramming is significantly affected by various biophysical factors, such as the nanostructure of the cell adhesion matrix, mechanical and electrical stimulations, pH, and oxygen tension, etc. Therefore, elements of the biophysical environment must be controlled in order to produce 3D cell aggregates, named embryoid bodies, in a bioreactor system (38-40). Oxygen concentration is a vital factor, by controlling pO2 at 30% air saturation, researchers have increased the growth rate, the energetic cell metabolism and the maximum cell concentration (41). Hydrodynamic shear stress is another factor that affects the differentiation efficiency. During the first 3 days, the differentiation protocol was adjusted by intermittent agitation, which resulted in more than 38% higher differentiation efficiency compared with static culture control (42). On this basis, the combination of hypoxia conditions with intermittent agitation gave rise to a 1000-fold increase in CM yields (43). By scaling up a multiwall screen to the bioreactor tank and applying intermittent agitation, researchers obtained 40 million predominantly ventricular-like CMs in 100 mL bioreactors with up to 85% purity (44). Furthermore, some studies have used Cytodex 3 as a microcarrier to differentiate hESCs in the bioreactor tank, and the yields were increased 12-fold compared to standard colony cultures by controlling the oxygen concentration at 20%. This scalable microcarrier system, from a few ml to more than 6,000 L of culture volumes using WNT signaling modulators, can be used to integrate cell propagation and differentiation. By optimizing differentiation conditions and controlling aggregate sizes, a yield of 1.9 million cells per ml was achieved with stirred spinner flask culturing (37). This combination of microcarriers that supports stem cell growth inside of a controlled stirred tank of bioreactors to provide stable differentiation processes hopefully becomes a more efficient solution to the large-scale expansion of hiPSCs. Electrical stimulus also facilitates the growth and differentiation of hiPSCs. By applying an electrical stimulus with frequencies between 0.033 and 1 Hz, cardiac contraction and relaxation were stimulated, and CM-specific genes could be detected (45,46).
Electrophysiological characteristics and various subtypes
HiPSCs are advantageous, as they provide a continuous source of cells by being reprogrammed from any somatic cells, such as skin fibroblasts, peripheral blood, extra-embryonic tissue from the umbilical cord and even urine (1,17,47-49). However, hiPSC-CMs are not identical to human CMs in terms of size, maturity, survival period, electrophysiological characteristics, and subtypes. HiPSC-CMs are considered immature when compared to mature CMs. They are smaller in size, resulting in a smaller maximum rate of potential depolarization, a smaller contractile force and a different impulse propagation. HiPSC-CMs are also round or multi-angular, usually single-nucleated, they do not present transverse tubules, and they have underdeveloped sarcoplasmic reticulum; whereas adult CMs are large and rod-like in shape, with 25–57% of the cells multi-nucleated (20,50). Moreover, like other cells with excitability, CMs are excited by the formation of local currents, and their electrophysiological characteristics are the main factors that determine and influence myocardial conductivity. The electrophysiological characteristics of hiPSCs are slightly different from those of mature CMs, and they also have different subtypes compared with CMs. In this section, we summarize the differences between the hiPSC-CMs and the CMs in vivo, in terms of electrophysiological characteristics and subtypes.
Various experiments have proven that iPSCs can be differentiated into functional CMs and respond. When exposed to β-adrenergic stimulation, an increasing spontaneous rate of reaction and a decreasing duration of action potential were detected (21,51-53). When analyzed by a multi-electrode assay, iPSC-CMs presented an identical dose-dependent change to the field potential waveform after applying ion channel inhibitors, compared to CMs. They were also capable of expressing specific cardiac markers, such as Nkx2.5, atrial natriuretic peptide, and GATA4 (5). However, there are still many differences that hinder the direct application of iPSC-CMs in clinical treatment. The CM clones derived from iPSCs that were generated from human foreskin fibroblasts were similar to certain parts of adult CMs in the excitation-contraction function. Nonetheless, the hiPSC-CMs exhibited a negative force-frequency relationship, milder post-rest potentiation, and less response to ryanodine and caffeine along with similar upstroke velocities with the human left ventricle tissue (53-55).
HiPSC-CMs have different survival periods compared with mature CMs. According to Shiba (56), after injection into a myocardial infarction model, hiPSC-CMs survived for 12 weeks and showed improved systolic function at weeks 4 and 12. Nonetheless, ventricular tachycardia was still a big barrier for clinical use, though it happened transiently. In the late stage (80–120 days), CMs had doubled contraction rates and slow contraction kinetics compared to early-stage CMs. They had hyperpolarized maximum diastolic potentials that increased the amplitudes of the action potentials and had faster upstroke velocities, suggesting that hiPSC-CMs mature slowly to reach a closer resemblance to adult CMs (57). However, the electrophysiological immaturity of hiPSC-CMs don’t preclude them from being accurate models for cardiac diseases. At least, the intact ion channel mechanisms of mature CMs also exist in hiPSC-CMs. In adult CMs, one important functional excitation-contraction coupling component is Ca2+. On one hand, the action potential triggers L-type Ca2+ channels, which further triggers a membrane Ca2+ current (58). On the other hand, the influx of Ca2+ triggers Ca2+-store release through Ca2+-sensitive ryanodine receptors, which are magnified several-fold by the sarohleasnic reticulum (58,59). In hiPSC-CMs, the whole-cell [Ca2+] transients depend on two pathways. One is the Ca2+ influx through L-type Ca2+ channels, and the other one is via intracellular stored Ca2+ release (60). The ionic currents and channel gating characteristics are based on their action potentials, which demonstrate the ability of hiPSC-CMs to be treated as models for functional gain and loss genetic disorders, thereby affecting the Na+ current (61,62).
Mature CMs can be divided into working CMs and cardiac pacemaker cells. Working CMs are the main part of the atrial and ventricular wall, and they are rich in myofibrils to have the conductivity, the excitability and the specific contractility necessary, while cardiac pacemaker cells have the same conductivity, excitability, and unique resultant rhythm. However, through recording action potentials and based on action potential characteristics, hiPSC-CMs have been recognized as having three categories: nodal cells, ventricular myocyte cells and atrial myocyte cells (Figure 2A). Those cardiac cells differ in regards to the composition of intracellular myofibril and the expression of ion channel proteins, which cause differences in action potential and contraction rhythm (Figure 2B) (63). Therefore, it is important to transplant a single subtype of CM according to the appropriate type and purity among the three subtypes of hiPSC-derived CMs. When done so, ventricular-like action potentials showed maximum diastolic potentials which are close to adult CMs (62).
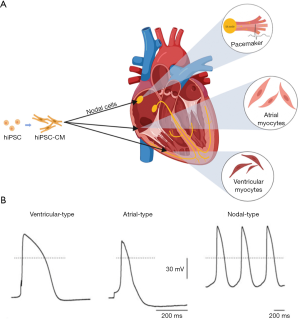
The expression of voltage-sensitive fluorescent proteins driven by subtype-specific promoters in atrial-, nodal-, and ventricular-like hiPSC-CMs allow different subtypes to be distinguished and allow precise disease phenotyping to be explored (64). Many methods have been developed to guide the differentiation towards an ideal subtype. When early clusters were transferred into an FBS-enriched culture medium, as soon as they were differentiated into spontaneously beating clusters, working-type cardiomyogenic differentiation was suppressed. The clusters were composed of 63.4% nodal-types and 36.6% atrial-types, while no ventricular-types were observed. These selectively differentiated cells with nodal-type properties were only media-based (65). In another research project, molecular beacons targeting mRNA related to working-type CMs were established to separate high throughput and specificity working-type CMs from nodal-type CMs (66). In addition, controlling the expansion in scalable suspension culture produced ventricular-like CMs with up to 85% purity (44).
Engineered 3D heart tissue using hiPSC-CMs
The main goal of tissue engineering is to build a functional tissue or organ which can reproduce the physical structure and physiological signals of the original tissue. To support these functions, cell-cell interaction is of great importance, especially between CMs and cardiac fibroblasts or endothelial cells. Cardiac fibroblasts in co-culture with CMs provide the appropriate mechanical properties to support the extracellular matrix, while endothelial cells provide advantages for essential nutrients and oxygen delivery, especially during the transition to thick tissues. In this section, we summarize the application of hiPSC-CMs in tissue engineering, including various appropriate biomaterials and microfabrication methods for achieving the regeneration and reproduction goal.
Biomaterials
Biomaterials provide environments where cells can adhere, survive, proliferate and differentiate. For creating functional 3D heart tissues, biomaterials should possess several unique properties such as biocompatibility, nanofibrous features, and biological activity to resemble native ECMs. Furthermore, the ability to tune the electrical conductivity and the mechanical properties of biomaterials could improve the maturation and differentiation of iPSC-CMs, making it possible to mimic the contractile behavior of native cardiac tissue. Several biocompatible materials, including natural polymer-based hydrogels, synthetic polymer based electrospun fibers, and decellularized native tissue have been applied to the creation of 3D heart tissue.
Hydrogels, including fibrin, gelatin, collagen, matrigel, chitosan and their various combinations, are the most common materials used for 3D bioprinting. Bioactive hydrogels such as gelatin, collagen and fibrin can support cell adhesion, cell encapsulation, oxygen and nutrient penetration, and secretion of waste products (67-70). The combination of different hydrogels such as gelatin-methacryloyl (GelMA) are reported to possess better performance in terms of cell stability and viability (71-74). In other studies, electrically conductive materials such as carbon nanotubes (CNT), gold nanoparticles, conducting polymers and reduced graphene oxide (rGO) were composited to GelMA or other biomaterials to improve electrical conductivity (75,76). Synthetic polymer based electrospun fibers provide stronger mechanical properties compared with hydrogels (77) and contribute to the formation of aligned cellular structures. Electrospun nanofibers from various materials, including poly (l-lactic acid) (PLA)/polyaniline (PANI) blend (78), PLA together with chitosan (79), gelatin (80), and silk fibroin/poly(ester-urethane) urea (81) have been used as scaffolds for heart tissue and have achieved positive results. On the other hand, decellularized native tissue made by removing xenogeneic cells from sacrificial tissue/organs can be a suitable scaffold for heart tissue since it retains the original extracellular matrix and membrane-associated proteins (82,83). By repopulating hiPSC-CMs to decellularized tissue, heart tissue with robust electrophysiological and contractile functions can be obtained (84,85). Each of the above biomaterials has advantages and drawbacks. Hydrogels are easy to fabric, store, and deliver, and are suitable for 3D bioprinting. However, they lack biological complexity and their mechanical strength is not able to pair with real heart tissue, especially in terms of anisotropy. Decellularized tissue almost perfectly duplicates real heart tissue, retaining the biological complexity, but is laborious to complete. Electrospun fibers provide favorable mechanical properties and conductivity but are hard to process into complex 3D structures.
Microfabrication methods
Native tissues and organs usually have well-organized and complex 3D structures consisting of various types of cells, extracellular matrices (ECM) and chemical and physical signaling cues (86,87). Particularly, in cardiac tissue, which consists of dense, quasi-lamellar and highly vascularized tissues, the functional syncytia of CMs tightly connects with gap junctions (88,89). In order to mimic the complex functions of native tissues, especially cardiac tissue, it is important to construct highly organized and functional 3D tissue structures in vitro, including various bioprinting and bioassembly approaches.
Bioprinting has emerged as a technology that can potentially address the above issues in the fabrication of vascularized cardiac tissue constructs, compared with conventional microfabrication techniques. Fabrication of artificial tissues or organs usually rely on additive manufacturing, also known as 3D printing. This method was firstly described in 1986 and later widely used in various areas such as engineering, manufacturing, and art. It consists of printing biocompatible materials, such as scaffolds, living cells and biochemicals into a complex artificial tissue, where they are precisely positioned into complex spatial structures to mimic the structure and function of living tissues. There are mainly three types of bioprinting methods: inkjet bioprinting, microextrusion, and photocuring bioprinting.
Inspired by conventional 2D inkjet printing, 3D inkjet bioprinting was the first technology applied to the microfabrication of artificial tissues (90). The extruded solution is stored in the ink cartridge and consists of biological materials, biochemicals, and living cells, which has also become known as a bioink. A substrate which can move relatively to the cartridge is placed below it to support the final construct (91,92). During printing, thermal (93) or acoustic force (92,94,95) is utilized to eject the bioink onto the substrate. The thermal ejection uses an electrical heater to heat the print head to produce vapor bubbles and generate air pressure which forces the droplet from the nuzzle. The acoustic ejection uses piezoelectric materials to generate jet force. When electrically actuated, these piezoelectric materials deform rapidly and press the liquid to eject droplets from the nozzle. The inkjet method can print at high speed and achieve relatively high cell viability (96-98). Meanwhile, the printing devices are usually affordable since they share a similar structure and machine components with commercial 3D printers. However, inkjet printers are not compatible with high viscosity or high cell density bioinks because of the low power of the heater or the piezoelectric actuator (99-102). Low viscosity bioinks can still settle in the cartridge, increasing the viscosity, therefore clogging the nozzle, which is known as the settling effect (101,102).
Extrusion printing is another common and affordable bioprinting method. Applying force through air pressure or mechanical screw allows the bioinks to extrude continuously and dispense themselves on the substrate layer by layer. This method is capable of printing almost all types of bioinks with various viscosities. However, extrusion printing has difficulty in precisely controlling the deposited mass. Additionally, extrusion printing is thought to reduce cell viability, since it applies a large mechanical stress on the cells (103,104). Photocuring bioprinting focuses UV light on the substrate to crosslink the photocurable polymer solution and scan over the surface to generate a layer as the substrate moves along the z-axis to enable 3D constructs. This technique allows for high resolution and thin layers, and can minimize potential cell damage by carefully optimizing the light source and the printing conditions. However, photocuring printing is only compatible with photocurable polymers, which limits the selection of bioinks. There are several other kinds of microfabrication methods for cardiac tissue, such as surface cell seeding on 3D printed scaffolds (105-107), creating channel networks within engineered tissue constructs by using sacrificial materials (108,109) and repopulating a decellularized matrix with a desired cell population (82,83,110). So far, no clinical cardiac tissue has been bioprinted; however, a combination of multiple bioprinting methods may further facilitate the development of the microfabrication of cardiac tissue.
Bioassembly refers to seeding cells in a predefined 2D or 3D construct, such as polymer scaffolds, ECM hydrogels or PDMS molds. Various bioassembly techniques were developed to facilitate cell aggregation, including acoustic field-guided assembly, magnetic field-guided assembly, gravity-driven assembly and molecular recognition-assisted self-assembly (111). In recent years, many techniques were applied to establishing 3D hiPSC-CMs tissues. For example, a cellular self-assembly cardiac tissue was developed by seeding hiPSC-CMs onto a frame assembled with a PDMS mold, which showed significantly different responses to drugs compared to monolayer cultured cells (112). Compared to 2D culturing, this formed tissue exhibited more functional cardiac tissue characteristics and had a more sensitive response to drugs with high throughput drug screening. Additionally, Faraday waves were used to promote hiPSC-CMs aggregated into predefined 3D constructs, rapidly enabling packing densities to resemble the architecture of the native myocardium at 108–109 cells/mL, which showed mature contractile function (113).
Recently, biomimetic scaffolds have been developed by optimizing the chitosan/collagen ratio and the temperature to achieve the stiffness of native cardiac tissue. The combination of directional micro-pores and a branched channel network enhanced the permeability of the scaffold. The synchronized beating of engineered myocardial tissue was achieved under electrical stimulation (114). Along with topographies, surface chemistry exhibited significant influence on the aggregation of hiPSC-CMs. Through aqueous atom transfer radical polymerization routes, poly (lactic acid) (PLA) microparticles were developed to mediate adhesion and enhanced contractility of hiPSC-CMs (115). Attempting to increase the efficiency and survival rate of CMs, bioassembly techniques avoid invasive fabrication and generate closely packed CMs-containing fabrication units through cell-driven organization in a short time.
Applications of iPSC-CMs
Heart failure has been a leading cause of death across the world over the last decade. Since myocardial cells are terminally differentiated with a limited capacity for division, and the adult heart lacks effective repair mechanisms, cell based regenerative therapy has been emerging as a novel therapeutic paradigm for myocardial repair. iPSC-CMs have been widely touted to have tremendous potential for repairing damaged heart tissue due to the large-scale production of hiPSC-CMs, providing a reliable source of cells for cardiac muscle transplants. In addition, there have been relatively few new drugs in the market for heart disease treatment (116,117). The main challenge in developing new drugs for heart disease is that the positive results from pre-clinical studies are often in contradiction with frequent frustrating outcomes in larger trials (116). A potential explanation for the challenges of developing new cardiac vascular disease (CVD) drugs is the lack of a suitable platform to screen for novel therapeutic strategies. In the last decade, drug toxicity and disease modeling have largely depended on animal models, which have serious drawbacks. Differences in pathophysiology, drug metabolism, and species-specific gene expression between humans and animals hinder the translational extension of testing the results of potential drugs’ efficacy and toxicity from animal models to clinical patients (9,116,117). HiPSCs can provide researchers with a limitless number of disease-relevant cells and can allow us to mimic the in vivo environment of a human. In the following parts, developments in the applications of iPSCs in regenerative medicine and drug screening are discussed in more detail.
Regenerative engineering
Now, there are two main effective strategies for treating heart failure, heart transplantation and ventricular assist devices (VADs). However, the development of current treatments for myocardial infarction (MI) are usually limited by many issues, including donor organ shortages, immunological rejection and invasive operations. Recently, the autologous transplantation of iPSC-CMs has had an extensive consensus to be a promising alternative strategy because of the exclusion of the possibility of immune rejection compared with other cell sources (118). The feasibility of autologous iPSC-CM therapy in clinical treatment is unclear because of the time, labor and cost, and the early-phase clinical trial results of autologous stem cell therapy are disappointing (118). Therefore, allogeneic transplantation of iPSC-CMs has attracted a lot of attention, despite the possibility of graft rejection. A potential approach for avoiding post-transplant immune response is by using major histocompatibility complex (MHC)-matched transplants. To fully validate the feasibility of MHC-matched allogeneic transplantation, Shiba et al. used a cynomolgus monkey whose MHC structure is similar to humans to establish an allogeneic transplantation model (56). Results showed that the iPSC-CMs injected directly into the intra-myocardial area successfully survived and that there was no evidence of immune rejection in monkeys subjected to myocardial infarction within 12 weeks. However, the incidence of non-lethal post-transplant ventricular tachycardia was observed, encouraging more efforts to investigate iPSC-CMs-induced arrhythmia before further studies in clinical settings. Besides the risk of serious arrhythmia, there are several major hurdles that need to be overcome before clinical application, such as the possibility of teratoma formation due to oncogenic factors from contaminating fractions of iPSCs, poor electrical uncoupling between grafted and native cardiac tissue, as well as the short survival time and retainment of implanted stem cells in the injured myocardium (56,118,119). To address these shortcomings, several approaches have been proposed. First, in terms of iPSC-CM purity, researchers genetically modified iPSCs in vitro during differentiation to express a Zeocin™ resistance gene in the presence of the cardiac-specific α-myosin heavy chain (α-MHC, MYH6) promoter (119). As shown in Figure 3A,B, transplanted purified iPSC-CM grafts developed typical cardiac features, e.g., mature sarcomeric structures, on the 7th day after intramyocardial injection. A study also reported that the inhibition of stearoyl-coA desaturase (SCD) could eliminate the tumorigenicity of undifferentiated iPSC derivatives by significantly decreasing the expression of Nanog, a marker for both cell pluripotency and tumor progression (121). Second, a comparable improvement of electrical coupling was also achieved by coculture with soluble factors secreted by MSCs, promoting the functional integration of iPSC-CMs into myocardial tissue (122). Third, to improve the survival and retention of transplanted iPSC-CMs, tissue engineering, an intelligent interdisciplinary strategy that combines cells, biomaterials and engineering methods to develop biological tissues, is becoming a promising strategy. Kawamura et al. introduced a cell-sheet method combined with a momentum flap to supply transplanted iPSC-CMs with enhanced blood via increased angiogenesis (123). Li et al. developed a folic acid derived hydrogel which could improve the survival and retention of encapsulated iPSC-CMs in MI models, improving the therapeutic efficacy (124). Masumoto et al. reported that they generated CMs, endothelial cells (ECs), and vascular mural cells (MCs) from hiPSCs and created engineered cardiac tissues (ECTs) composed of these three cellular compositions, validating that the incorporation of vascular cells derived from hiPSCs could enhance the maturation and function of cardiac tissue (125). Regarding engineering methods, it must be mentioned that Noor et al. developed a simple bioprinting approach to fabricate a vascularized cardiac patch using iPSC-CMs and EC. The capability to print a functional vascularized patch according to patient’s anatomy further validates the potential of heart engineering in clinical regenerative therapy (126).
The delivery of cardiac tissue derived from hiPSC-CMs has demonstrated a promising therapeutic strategy for cardiac repair after MI. Although the efforts mentioned above have decreased the clinical risk, their effects and mechanisms of action remain unclear. Recently, some studies described a new cell free therapy that could improve cardiac recovery after MI through the delivery of exosomes isolated from iPSC-CMs (127-129). The direct injection of extracellular vesicles (EV) secreted by iPSC-CMs into MI hearts has shown a strong repairing effect in preclinical animal models (127). The delivery of EVs from cardiac patches and hydrogels to the heart in a sustainable and minimally invasive manner could be realized thanks to advances in biomaterials. Reporting in Nature Biomedical Engineering, Gordana Vunjak-Novakovic and colleagues found that, compared with iPSC-EVs, iPSC-CM-EVs have greater therapeutic potential in vitro. They encapsulated iPSC-CM-EVs into a collagen gel foam mesh to develop a hydrogel patch capable of delivering EVs continuously (128). They successfully proved that the delivery of EVs from the hydrogel patch attenuated pathological hypertrophy and promoted functional recovery after ischemic injury. Despite more effort being needed to completely understand the underlying mechanisms behind the function of iPSC-CM-EVs, this cell-free therapy based on EVs represents a brand-new direction for heart recovery investigation and heart injury treatment.
In vitro models
The use of iPSCs in cardiovascular research has attracted a lot of attention due to iPSC-CMs having similar properties to CMs in vitro, including numerous drug-affected targets and disease-related genetic mutations, making it possible to use these cells for drug screening with or without inducing specific disease phenotypes, as well as exploring the pathological mechanisms of heart disease (130).
Without specific induced phenotypes, iPSC-CMs can be used as a pre-clinical cardiac platform for screening the cardiotoxicity of drug candidates before clinical testing (14,130-138). Particularly, rare drug-induced fatal ventricular arrhythmia, such as torsades de pointes (TdP), is the focus of cardiac safety within the current regulatory guidelines (14,139). By evaluating the current state of drug-induced TdP iPSC-CM models through a microelectrode array (MEA) and voltage-sensing optical (VSO) techniques, Blinova et al. recently used 2 commercial hiPSC-CMs to detect the cellular electrophysiology (EP) effect of 28 drugs which have been known to have clinical TdP risk (117). However, several experimental limitations of hiPSC-CM assays in TdP risk are worth noting. This study is not statistically significant enough due to the limitations of cell sources, experimental sites, readouts and culture conditions, especially the fact that hiPSCs from healthy people may differ significantly from patients in terms of genetic predisposition, which decide the personal clinical risk of arrhythmia (117,140). Moreover, the cardiotoxicity effects or cardioprotective responses of new chemical entities (NCEs) are variable between different iPSC lines, which can be explained by the genetic differences of the donors. The clinical cardiotoxicity of chemotherapeutic agents varies largely among different people, from 8% to 26% for doxorubicin, 5% to 30% for paclitaxel, or 7% to 28% for trastuzumab (117,140). Considering the genetic diversity of clinical patients, the sources of iPSC-CMs should be enlarged. Choosing donors with varied backgrounds might improve the predictivity of the iPSC-CM cardiac safety screening platform by taking a lot of factors into consideration, such as gender, race, and genetic diversity.
In the case of disease-specific iPSC-CM models, Drawnel et al. used iPSC-CMs to create environmental in vitro models of diabetic cardiomyopathy (DCM) by exposing cells to a diabetogenic environment so that the cells could recapitulate a phenotypic surrogate of DCM (141). Through stimulating commercial iPSC-CMs with hydrogen peroxide, Fiedler et al. developed an ischemic injury model characterized by a loss of CMs that damages heart function, which is the most probable pathological mechanism of heart failure (137). These studies confirmed the utility of hiPSC-CM models in drug discovery to validate targets and develop compounds for target treatments. Furthermore, using hiPSC-CMs inherited from diseased individuals, patient specific-disease models could be applied to a drug screening platform that could test drug candidates and identify disease targets for the discovery of therapeutic strategies for diseases.
In addition to cardiac disease models, iPSC-CMs are potential platforms for drug toxicity models, especially chemotherapy-induced cardiotoxicity. Cardiac dysfunction induced by chemotherapies could be recapitulated by hiPSC-CMs at a single-cell level. For example, Burridge et al. generated hiPSC-CMs from breast cancer patients with clinical doxorubicin cardiotoxicity and found that these cells had a predilection for doxorubicin side effects (142). The in vitro cardiac model presented numerous doxorubicin-induced cardiotoxicity phenotypes, such as sarcomere disruption, oxidative stress, and CM death. One recent study found that iPSC-CMs generated from patients receiving trastuzumab therapy could recapitulate the patient-specific response to trastuzumab therapy and the clinical features of patients’ phenotypes in vitro (10). They also demonstrated the therapeutic potential of targeting the altered metabolism pathway, which underlies the cardiac dysfunction induced by trastuzumab therapy, in mitigating the side effects of trastuzumab therapy in patients with cancer. Taken together, drug toxicity models enable the investigation of the mechanisms of drug-induced cardiac dysfunction and the ability to search for a potential therapy for a specific target.
Hearts-on-chips
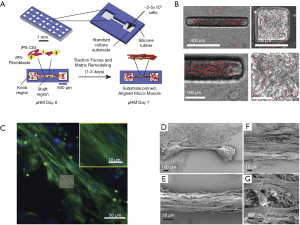
Heart-on-chips, also known as micro physiological systems (MPS), have emerged as a promising candidate for cardiac safety and for the efficacy of screening platforms for preclinical NCEs. The heart-on-a-chip is an innovative multidiscipline 3D cardiac tissue fabrication approach to reproduce cardiac tissue for the study of the heart (11,117,143,144). Microsystem engineering allows researchers to develop a micro-physiological environment composed of cardiac cells that recapitulate some level of cardiac tissue or organ architecture and function in vitro with the advantage needing fewer cells and having higher reproducibility (144). Mathur et al. successfully developed a cardiac microphysiological system to predict the cardiotoxicity of drugs. This in vitro platform showed a dose-dependent toxicity of several drugs on the cardiac constructs which was consistent with clinical observations (Figure 3C,D,E,F) (120). Weng et al. also developed a 3D microfluidic device comprised of tumor cell spheroids, iPSC-CMs and iPS-ECs. This iPSC-CM-EC-tumor-on-a-chip could simultaneously assess cardiac toxicity and the anti-tumor effects of drugs at the same time (145). The iPSC-CM based heart-on-a-chip is widely recognized as a miniaturized 3D cardiac tissue and heart model. However, before the iPSC-CM based heart-on-a-chip becomes a useful approach for drug discovery in the industry, there are still some challenges that need to be addressed beforehand, such as a lack of large-scale production, a continuous and automated process, as well as quality control in the fabrication process. To solve these technical problems, many novel strategies were explored by researchers. Lind et al. developed an easy-handle approach to fabricate a new class of iPSC-CM based heart-on-a-chip using 3D printing. By designing six functional inks, they obtained physio-mimetic cardiac tissue and intact integrated sensors. The drug response of this heart-on-a-chip was validated by assessing the contractile profile of cardiac tissue over one month (146). Additionally, the predictive power of this heart-on-a-chip for pharmaceutical screening was significantly affected by the functional assembly and maturation of iPSC-CMs. Considering the impact of pericellular micro-environment cues on individual iPSC-CMs, Zhao et al. investigated the effects of cell seeding density, cell types, the percentage of non-myocyte populations, and the types of hydrogels used for tissue inoculation and electrical conditioning regimes on iPSC-CM assembly using a novel heart-on-a-chip system. They confirmed that the robustness and fidelity of iPSC-CM-derived cardiac tissue for drug screening and disease modeling could be improved by optimizing these micro-environmental cues. It was also indicated that the heart-on-a-chip system could be used as an in vitro platform for exploring the mechanisms underlying iPSC-CM differentiation and maturation (147). Huebsch et al. (148) also developed a new approach that combined features of engineered heart muscle and cardiospheres to produce miniaturized heart-on-chips: Micro-Heart Muscle (µHM) arrays, which were presented as a new platform for studying CM maturation, disease and cardiotoxicity in vitro (Figure 4A). After optimizing the design criteria for each individual component of the dogbone-shape array, uniaxial beating was successfully induced in a 100 µm width channel in the shaft region (Figure 4B). It was further found that the sarcomeres of µHM formed within heart-on-chips exhibited extensive organization throughout the shaft region (Figure 4C). Additionally, morphologically complex fibers of µHM were shown in SEM images (Figure 4D,E,F,G). In this way, a simple fabrication strategy to yield superior morphological and functional µHMs using a small quantity of iPSC-CMs was presented, exploring the application of iPSC-CMs in disease modeling and drug screening (148).
The tissue composition and construction of an in vitro 3D microdevice incorporated with vascular perfusion could be precisely controlled with microfabrication techniques. To reproduce the hierarchical structure of the native myocardium, Zhang et al. proposed an innovative 3D bioprinting technology-based hybrid strategy to fabricate a myocardium integrated with blood vessels (149). They directly printed a 3D construct using a microfibrous hydrogel encapsulated with endothelial cells that could form a confluent endothelium, followed by seeding CMs derived from iPSC-CMs that could generate an aligned myocardium. Then, this multi-cell 3D bed was embedded into a bioreactor with microfluidic perfusion to create a composite endothelium-myocardium-on-a-chip platform. The heart-on-a-chip could be applied to preclinical drug evaluation and regenerative medicine as a highly promising study platform for drug discovery and for modeling inheritable cardiac disease (11,130,150).
Future uses of iPSC-CMs in drug screening: personalized medicine
Clinically, genetic differences among patients that may contribute to personalized cardiac phenotypes and pathology mechanisms have greatly hindered the drug discovery process and have reduced treatment efficacy (65,66). Therefore, hiPSC-CMs have emerged as an exciting paradigm to revolutionize the discovery of new drugs for serious heart conditions because they are a patient-specific cell source and because of their ability to generate heart tissue models which can be used for personalized drug screening platforms and for understanding patient-specific disease mechanisms (151-153). These “trials in-a-dish” have gained popularity among many researchers who are now using iPSC-CM models to predict both the cardiac toxicity and the treatment effectiveness of drugs, as well as for testing therapeutic strategies (10,116,154). In the future, it is conceivable that an iPSC technology-based screening platform might make personalized “drug screens” possible at an individual level (117,142,155). By generating iPSCs from a patient’s somatic cells, derived CMs from patient-specific iPSCs were used to represent single-cell phenotype features of a specific patient and their disease (120,156). In the study mentioned above, Drawnel et al. found out that diabetic patient-derived iPSC-CMs showed DCM phenotypes, which depend on the clinical status of the original donor, without exposure to diabetic stimulus, showing the potential of patient-derived iPSC-CMs for pathogenesis research and drug discovery (141). The recapitulation of the DCM phenotype at different clinical statuses in patient specific iPSC-CMs was demonstrated in many aspects, such as CM score, frequency of calcium transients, irregularity of beating rate, levels of intracellular lipid and the extent of lipid peroxidation. Consequently, it should be possible to develop personalized drugs for specific individual targets that are unique for specific groups of patients, thereby improving treatment efficiency and reducing adverse effects in clinical practice. Briefly, the hiPSC-CMs facilitate the development of personalized medicine approaches, which depend on the response of patient-specific cells to drugs (141).
Challenges and future perspectives
The manufacturing of human iPSC master cell banks has been reported recently and the cells have proven to differentiate towards specific lineages, which represents a significant breakthrough in the field of regenerative medicine. Potential applications of iPSC-CMs include cardiac regeneration therapy, drug selection and disease models. Since 1997, the number of drugs withdrawn from clinical use keeps increasing due to cardiotoxicity (157). Nowadays, high-throughput drug cardiotoxicity screening based on hiPSC-CMs has gradually matured, which will greatly reduce the research and development costs. However, the efficiency and economy of the large-scale producing methods for hiPSC-CMs still need to be improved. Models that elucidate the genetic and epigenetic mechanisms that regulate cell differentiation will be of great help to the large-scale production and purification of iPSC-CMs. Other challenges which surround iPSC-CMs also include tumorigenesis in long-term culture and the low level of iPSC-CMs’ maturity.
Despite advances in exploring strategies for regeneration therapy, including 3D printing, organ-on-a-chip, and the molecular basis of artificial cardiac tissue, their roles in CM differentiation, the function of mature specific subtype CMs and disease development remain rarely defined. A major impedance is the immaturity of hiPSC-CMs in contractility, calcium handling and electrophysiology. The main reason is that specific environmental stimuli and transcriptional regulations are required to synergistically trigger maturation (158). Additionally, there are no fully accepted criteria to assess hiPSCs, whether in transcriptomic, genetic or epigenetic approaches.
A potential technical obstruction of the application of iPSC-CMs is the obvious immature phenotype in vitro differentiated iPSC-CMs have in terms of ultrastructure and electrophysiological properties. Indeed, the cellular immaturity of iPSC-CMs was exemplified in one study mentioned above. Paul W. Burridge et al. evaluated the effect of a cardioprotective compound in the presence of doxorubicin on iPSC-CMs, in which dexrazoxane, the drug against doxorubicin-induced cardiotoxicity, showed increased toxicity, diverging from the clinical experience (142). Several methods have been proposed to promote CM maturation in vitro, including growth factors, electrical and mechanical stimulation, and 3D culture (158-163). To some extent, the immaturity of iPSC-CMs could be explained by the lack of information on cell-cell and cell-matrix communication in a 2D monolayer culture platform. In addition, it is worth mentioning that the responses of CMs to injury and drugs are dramatically affected by the native heart environment, which is composed of cardiac fibroblasts, vascular endothelial cells, smooth muscle cells, leukocytes and an organized extracellular matrix (9,17,154,164). Furthermore, in contrast to electrophysiological disorders and cell-autonomous disorders, noncellular autonomic pathogenesis and structure-related disease phenotypes are more closely correlated to other types of cells in heart tissues other than CMs. Compared with the traditional two-dimensional (2D) monolayer culture method, 3D iPSC-CM cardiac models, when integrated with other cardiac cell types, are more identical to the in vivo environment of human clinical pathophysiology than 2D models. The 3D model could provide both a biomimetic and complex environment to mimic in vivo. In such a case, there is a compelling need for further refinements to construct a 3D heart model to mimic the in vivo heart environment faithfully. But there have been big issues for the therapeutic strategy using iPSCs. The strategy cannot be performed quickly enough to treat patients with their specific somatic cells, because many CVDs progress rapidly while many aspects of establishing iPSCs take long periods of time, such as reprograming cells, maintaining or expanding cell numbers and ensuring the quality and safety of the procedure. That is the reason why the consortium was built by Dr. Shinya Yamanaka et al. (165) who developed iPSCs. The consortium is gathering somatic cells from healthy human leukocyte antigens (HLA) homo donors with around 100–300 different HLA. Those homo HLA cells are stocked after reprograming them into iPSCs and after qualification. They cover more than 90% of the popularity in the world and can be directly transplanted into patients with the same HLA type (165).
Conclusions
HiPSCs are the only reliable source of CMs and cardiac progenitor cells at present, offering hope for clinical applications in patients who need cardiac regeneration therapy. HiPSC-CMs avoid interspecies differences and function as an efficient drug screening platform for humans while enhancing the development of personalized medicine. Methods for inducing iPSC differentiation and for purifying iPSC-CMs have been developed, as well as engineering approaches that allow for the large-scale production of iPSC-CMs. Several new generations of strategies to strengthen the possibilities of iPSC-CM applications for clinical use have been proposed, including adding miRNA or micro molecules, improving 3D scaffolds, establishing 3D iPSC-CM cardiac models integrated with various types of CMs and building specific iPSC technology-based screening platforms. However, these studies are still in their early stages and have not yet been devoted to clinical use. Researchers still need to make efforts in many aspects, such as avoiding inter batch variation and arrhythmia, improving the long-term viability and the cell purification, and predicting which maturity level of iPSC-CMs are better to be transplanted in order to functionally integrate with host CMs. Nevertheless, it is clear that in the future, regenerative medicine will become more efficient and accurate, and iPSC-CMs will play an important role in screening cardiac drug development and promoting tissue repair.
Acknowledgments
Funding: This paper was funded by AHA Innovative Project Award (19IPLOI34660079), Qatar University grants NPRP9-144-3-021 and NPRP12 NPRP12S-0310-190276 funded by Qatar National Research Foundation, a part of Qatar Foundation, and the Brigham Research Institute Stepping Strong Innovator Award and Innovation Evergreen Fund.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/mps-20-3). The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007;131:861-72. [Crossref] [PubMed]
- Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007;318:1917-20. [Crossref] [PubMed]
- Moretti A, Bellin M, Welling A, et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med 2010;363:1397-409. [Crossref] [PubMed]
- Ye L, Chang YH, Xiong Q, et al. Cardiac repair in a porcine model of acute myocardial infarction with human induced pluripotent stem cell-derived cardiovascular cells. Cell stem cell 2014;15:750-61. [Crossref] [PubMed]
- Tanaka T, Tohyama S, Murata M, et al. In vitro pharmacologic testing using human induced pluripotent stem cell-derived cardiomyocytes. Biochem Biophys Res Commun 2009;385:497-502. [Crossref] [PubMed]
- Liang P, Lan F, Lee AS, et al. Drug screening using a library of human induced pluripotent stem cell–derived cardiomyocytes reveals disease-specific patterns of cardiotoxicity. Circulation 2013;127:1677-91. [Crossref] [PubMed]
- Huang CY, Liu CL, Ting CY, et al. Human iPSC banking: barriers and opportunities. J Biomed Sci 2019;26:87. [Crossref] [PubMed]
- Liang P, Sallam K, Wu H, et al. Patient-specific and genome-edited induced pluripotent stem cell–derived cardiomyocytes elucidate single-cell phenotype of Brugada syndrome. J Am Coll Cardiol 2016;68:2086-96. [Crossref] [PubMed]
- Sayed N, Ameen M, Wu JC. Personalized medicine in cardio-oncology: the role of induced pluripotent stem cell. Cardiovasc Res 2019;115:949-59. [Crossref] [PubMed]
- Kitani T, Ong S-G, Lam CK, et al. Human-induced pluripotent stem cell model of trastuzumab-induced cardiac dysfunction in patients with breast cancer. Circulation 2019;139:2451-65. [Crossref] [PubMed]
- Ramme AP, Koenig L, Hasenberg T, et al. Towards an autologous iPSC-derived patient-on-a-chip. bioRxiv 2018:376970.
- Eschenhagen T, Mummery C, Knollmann BC. Modelling sarcomeric cardiomyopathies in the dish: from human heart samples to iPSC cardiomyocytes. Cardiovasc Res 2015;105:424-38. [Crossref] [PubMed]
- Malik N, Rao MS. A review of the methods for human iPSC derivation. Methods Mol Biol 2013;997:23-33. [Crossref] [PubMed]
- Karakikes I, Ameen M, Termglinchan V, et al. Human induced pluripotent stem cell–derived cardiomyocytes: insights into molecular, cellular, and functional phenotypes. Circ Res 2015;117:80-8. [Crossref] [PubMed]
- Masumoto H, Yamashita JK. Human iPS cell-derived cardiac tissue sheets: a platform for cardiac regeneration. Curr Treat Options Cardiovasc Med 2016;18:65. [Crossref] [PubMed]
- Park M, Yoon YS. Cardiac regeneration with human pluripotent stem cell-derived cardiomyocytes. Korean Circ J 2018;48:974-88. [Crossref] [PubMed]
- Smith AS, Macadangdang J, Leung W, et al. Human iPSC-derived cardiomyocytes and tissue engineering strategies for disease modeling and drug screening. Biotechnol Adv 2017;35:77-94. [Crossref] [PubMed]
- Del Álamo JC, Lemons D, Serrano R, et al. High throughput physiological screening of iPSC-derived cardiomyocytes for drug development. Biochim Biophys Acta 2016;1863:1717-27. [Crossref] [PubMed]
- Batalov I, Feinberg AW. Differentiation of Cardiomyocytes from Human Pluripotent Stem Cells Using Monolayer Culture. Biomark Insights 2015;10:71-6. [PubMed]
- Robertson C, Tran DD, George SC. Concise review: Maturation phases of human pluripotent stem cell-derived cardiomyocytes. Stem cells 2013;31:829-37. [Crossref] [PubMed]
- Zhang J, Wilson GF, Soerens AG, et al. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res 2009;104:e30-41. [Crossref] [PubMed]
- Zwi L, Caspi O, Arbel G, et al. Cardiomyocyte differentiation of human induced pluripotent stem cells. Circulation 2009;120:1513. [Crossref] [PubMed]
- Kawamura M, Miyagawa S, Miki K, et al. Feasibility, safety, and therapeutic efficacy of human induced pluripotent stem cell-derived cardiomyocyte sheets in a porcine ischemic cardiomyopathy model. Circulation 2012;126:S29-37. [Crossref] [PubMed]
- Olson EN, Schneider MD. Sizing up the heart: development redux in disease. Genes Dev 2003;17:1937-56. [Crossref] [PubMed]
- Elliott DA, Braam SR, Koutsis K, et al. NKX2-5 eGFP/w hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat Methods 2011;8:1037. [Crossref] [PubMed]
- Kubo A, Shinozaki K, Shannon JM, et al. Development of definitive endoderm from embryonic stem cells in culture. Development 2004;131:1651-62. [Crossref] [PubMed]
- Ueno S, Weidinger G, Osugi T, et al. Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc Natl Acad Sci U S A 2007;104:9685-90. [Crossref] [PubMed]
- Burridge PW, Thompson S, Millrod MA, et al. A universal system for highly efficient cardiac differentiation of human induced pluripotent stem cells that eliminates interline variability. PLoS One 2011;6:e18293 [Crossref] [PubMed]
- Ding VM, Ling L, Natarajan S, et al. FGF-2 modulates Wnt signaling in undifferentiated hESC and iPS cells through activated PI3-K/GSK3β signaling. J Cell Physiol 2010;225:417-28. [Crossref] [PubMed]
- Mummery C, Ward-van Oostwaard D, Doevendans P, et al. Differentiation of human embryonic stem cells to cardiomyocytes: role of coculture with visceral endoderm-like cells. Circulation 2003;107:2733-40. [Crossref] [PubMed]
- Passier R, Oostwaard DW, Snapper J, et al. Increased cardiomyocyte differentiation from human embryonic stem cells in serum-free cultures. Stem Cells 2005;23:772-80. [Crossref] [PubMed]
- Burridge PW, Matsa E, Shukla P, et al. Chemically defined generation of human cardiomyocytes. Nat Methods 2014;11:855-860. [Crossref] [PubMed]
- Yang L, Soonpaa MH, Adler ED, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature 2008;453:524-8. [Crossref] [PubMed]
- Zhang J, Klos M, Wilson GF, et al. Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: the matrix sandwich method. Circ Res 2012;111:1125-36. [Crossref] [PubMed]
- Lian X, Zhang J, Azarin SM, et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nature protocols 2013;8:162. [Crossref] [PubMed]
- Minami I, Yamada K, Otsuji TG, et al. A small molecule that promotes cardiac differentiation of human pluripotent stem cells under defined, cytokine-and xeno-free conditions. Cell Rep 2012;2:1448-60. [Crossref] [PubMed]
- Lemoine MD, Mannhardt I, Breckwoldt K, et al. Human iPSC-derived cardiomyocytes cultured in 3D engineered heart tissue show physiological upstroke velocity and sodium current density. Sci Rep 2017;7:5464. [Crossref] [PubMed]
- Forsyth NR, Musio A, Vezzoni P, et al. Physiologic oxygen enhances human embryonic stem cell clonal recovery and reduces chromosomal abnormalities. Cloning Stem Cells 2006;8:16-23. [Crossref] [PubMed]
- Gibbons J, Hewitt E, Gardner D. Effects of oxygen tension on the establishment and lactate dehydrogenase activity of murine embryonic stem cells. Cloning Stem Cells 2006;8:117-22. [Crossref] [PubMed]
- Bauwens C, Yin T, Dang S, et al. Development of a perfusion fed bioreactor for embryonic stem cell-derived cardiomyocyte generation: Oxygen-mediated enhancement of cardiomyocyte output. Biotechnol Bioeng 2005;90:452-61. [Crossref] [PubMed]
- Serra M, Brito C, Sousa MF, et al. Improving expansion of pluripotent human embryonic stem cells in perfused bioreactors through oxygen control. J Biotechnol 2010;148:208-15. [Crossref] [PubMed]
- Ting S, Chen A, Reuveny S, et al. An intermittent rocking platform for integrated expansion and differentiation of human pluripotent stem cells to cardiomyocytes in suspended microcarrier cultures. Stem Cell Res 2014;13:202-13. [Crossref] [PubMed]
- Correia C, Serra M, Espinha N, et al. Combining hypoxia and bioreactor hydrodynamics boosts induced pluripotent stem cell differentiation towards cardiomyocytes. Stem Cell Rev Rep 2014;10:786-801. [Crossref] [PubMed]
- Kempf H, Olmer R, Kropp C, et al. Controlling expansion and cardiomyogenic differentiation of human pluripotent stem cells in scalable suspension culture. Stem Cell Reports 2014;3:1132-46. [Crossref] [PubMed]
- Ge D, Liu X, Li L, et al. Chemical and physical stimuli induce cardiomyocyte differentiation from stem cells. Biochem Biophys Res Commun 2009;381:317-21. [Crossref] [PubMed]
- Geuss LR, Suggs LJ. Making cardiomyocytes: how mechanical stimulation can influence differentiation of pluripotent stem cells. Biotechnol Prog 2013;29:1089-96. [Crossref] [PubMed]
- Loh YH, Agarwal S, Park IH, et al. Generation of induced pluripotent stem cells from human blood. Blood 2009;113:5476-9. [Crossref] [PubMed]
- Cai J, Li W, Su H, et al. Generation of human induced pluripotent stem cells from umbilical cord matrix and amniotic membrane mesenchymal cells. J Biol Chem 2010;285:11227-34. [Crossref] [PubMed]
- Xue Y, Cai X, Wang L, et al. Generating a non-integrating human induced pluripotent stem cell bank from urine-derived cells. PLoS One 2013;8:e70573 [Crossref] [PubMed]
- Pekkanen-Mattila M, Häkli M, Pölönen RP, et al. Polyethylene Terephthalate Textiles Enhance the Structural Maturation of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Materials 2019;12:1805. [Crossref] [PubMed]
- Bylund JB, Trinh LT, Awgulewitsch CP, et al. Coordinated proliferation and differentiation of human-induced pluripotent stem cell-derived cardiac progenitor cells depend on bone morphogenetic protein signaling regulation by GREMLIN 2. Stem Cells Dev 2017;26:678-93. [Crossref] [PubMed]
- Chai S, Wan X, Nassal DM, et al. Contribution of two-pore K+ channels to cardiac ventricular action potential revealed using human iPSC-derived cardiomyocytes. Am J Physiol Heart Circ Physiol 2017;312:H1144-53. [Crossref] [PubMed]
- Germanguz I, Sedan O, Zeevi-Levin N, et al. Molecular characterization and functional properties of cardiomyocytes derived from human inducible pluripotent stem cells. J Cell Mol Med 2011;15:38-51. [Crossref] [PubMed]
- Branco MA, Cotovio JP, Rodrigues CAV, et al. Transcriptomic analysis of 3D Cardiac Differentiation of Human Induced Pluripotent Stem Cells Reveals Faster Cardiomyocyte Maturation Compared to 2D Culture. Sci Rep 2019;9:9229. [Crossref] [PubMed]
- Pieske B, Kretschmann B, Meyer M, et al. Alterations in intracellular calcium handling associated with the inverse force-frequency relation in human dilated cardiomyopathy. Circulation 1995;92:1169-78. [Crossref] [PubMed]
- Shiba Y, Gomibuchi T, Seto T, et al. Allogeneic transplantation of iPS cell-derived cardiomyocytes regenerates primate hearts. Nature 2016;538:388-91. [Crossref] [PubMed]
- Lundy SD, Zhu W-Z, Regnier M, et al. Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cells Dev 2013;22:1991-2002. [Crossref] [PubMed]
- Bers DM. Cardiac excitation–contraction coupling. Nature 2002;415:198-205. [Crossref] [PubMed]
- Cannell MB, Cheng H, Lederer WJ. The control of calcium release in heart muscle. Science 1995;268:1045-9. [Crossref] [PubMed]
- Itzhaki I, Rapoport S, Huber I, et al. Calcium handling in human induced pluripotent stem cell derived cardiomyocytes. PLoS One 2011;6:e18037 [Crossref] [PubMed]
- Davis RP, Casini S, van den Berg CW, et al. Cardiomyocytes derived from pluripotent stem cells recapitulate electrophysiological characteristics of an overlap syndrome of cardiac sodium channel disease. Circulation 2012;125:3079-91. [Crossref] [PubMed]
- Ma J, Guo L, Fiene SJ, et al. High purity human-induced pluripotent stem cell-derived cardiomyocytes: electrophysiological properties of action potentials and ionic currents. Am J Physiol Heart Circ Physiol 2011;301:H2006-17. [Crossref] [PubMed]
- Lin Y, Liu H, Klein M, et al. Efficient differentiation of cardiomyocytes and generation of calcium-sensor reporter lines from nonhuman primate iPSCs. Sci Rep 2018;8:5907-16. [Crossref] [PubMed]
- Chen Z, Xian W, Bellin M, et al. Subtype-specific promoter-driven action potential imaging for precise disease modelling and drug testing in hiPSC-derived cardiomyocytes. Eur Heart J 2017;38:292-301. [PubMed]
- Schweizer PA, Darche FF, Ullrich ND, et al. Subtype-specific differentiation of cardiac pacemaker cell clusters from human induced pluripotent stem cells. Stem Cell Res Ther 2017;8:229. [Crossref] [PubMed]
- Jha R, Wile B, Wu Q, et al. Molecular beacon-based detection and isolation of working-type cardiomyocytes derived from human pluripotent stem cells. Biomaterials 2015;50:176-85. [Crossref] [PubMed]
- Bertassoni LE, Cardoso JC, Manoharan V, et al. Direct-write bioprinting of cell-laden methacrylated gelatin hydrogels. Biofabrication 2014;6:024105 [Crossref] [PubMed]
- Billiet T, Gevaert E, De Schryver T, et al. The 3D printing of gelatin methacrylamide cell-laden tissue-engineered constructs with high cell viability. Biomaterials 2014;35:49-62. [Crossref] [PubMed]
- Loo Y, Lakshmanan A, Ni M, et al. Peptide bioink: self-assembling nanofibrous scaffolds for three-dimensional organotypic cultures. Nano letters 2015;15:6919-25. [Crossref] [PubMed]
- Ozbolat IT, Hospodiuk M. Current advances and future perspectives in extrusion-based bioprinting. Biomaterials 2016;76:321-43. [Crossref] [PubMed]
- Yue K, Trujillo-de Santiago G, Alvarez MM, et al. Synthesis, properties, and biomedical applications of gelatin methacryloyl (GelMA) hydrogels. Biomaterials 2015;73:254-71. [Crossref] [PubMed]
- Klotz BJ, Gawlitta D, Rosenberg AJ, et al. Gelatin-methacryloyl hydrogels: towards biofabrication-based tissue repair. Trends Biotechnol 2016;34:394-407. [Crossref] [PubMed]
- Colosi C, Shin SR, Manoharan V, et al. Microfluidic bioprinting of heterogeneous 3D tissue constructs using low-viscosity bioink. Adv Mater 2016;28:677-84. [Crossref] [PubMed]
- Liu W, Heinrich MA, Zhou Y, et al. Extrusion bioprinting of shear-thinning gelatin methacryloyl bioinks. Adv Healthc Mater 2017;6:1601451 [Crossref] [PubMed]
- Shin SR, Zihlmann C, Akbari M, et al. Reduced graphene oxide-gelMA hybrid hydrogels as scaffolds for cardiac tissue engineering. Small 2016;12:3677-89. [Crossref] [PubMed]
- Shin SR, Jung SM, Zalabany M, et al. Carbon-nanotube-embedded hydrogel sheets for engineering cardiac constructs and bioactuators. ACS Nano 2013;7:2369-80. [Crossref] [PubMed]
- Kaneko N, Matsuda R, Toda M, et al. Three-dimensional reconstruction of the human capillary network and the intramyocardial micronecrosis. Am J Physiol Heart Circ Physiol 2011;300:H754-61. [Crossref] [PubMed]
- Wang L, Wu Y, Hu T, et al. Electrospun conductive nanofibrous scaffolds for engineering cardiac tissue and 3D bioactuators. Acta Biomater 2017;59:68-81. [Crossref] [PubMed]
- Liu Y, Wang S, Zhang R. Composite poly (lactic acid)/chitosan nanofibrous scaffolds for cardiac tissue engineering. Int J Biol Macromol 2017;103:1130-7. [Crossref] [PubMed]
- Elamparithi A, Punnoose AM, Paul SF, et al. Gelatin electrospun nanofibrous matrices for cardiac tissue engineering applications. Int J Polym Mater Polym Biomater 2017;66:20-7. [Crossref]
- Du J, Zhu T, Yu H, et al. Potential applications of three-dimensional structure of silk fibroin/poly (ester-urethane) urea nanofibrous scaffold in heart valve tissue engineering. Appl Surf Sci 2018;447:269-78. [Crossref]
- Dohmen PM, Scheckel M, Stein-Konertz M, et al. In vitro hydrodynamics of a decellularized pulmonary porcine valve, compared with a glutaraldehyde and polyurethane heart valve. Int J Artif Organs 2002;25:1089-94. [Crossref] [PubMed]
- Juthier F, Vincentelli A, Gaudric J, et al. Decellularized heart valve as a scaffold for in vivo recellularization: deleterious effects of granulocyte colony-stimulating factor. J Thorac Cardiovasc Surg 2006;131:843-52. [Crossref] [PubMed]
- Garreta E, De Oñate L, Fernández-Santos ME, et al. Myocardial commitment from human pluripotent stem cells: Rapid production of human heart grafts. Biomaterials 2016;98:64-78. [Crossref] [PubMed]
- Guyette JP, Charest JM, Mills RW, et al. Bioengineering human myocardium on native extracellular matrix. Circ Res 2016;118:56-72. [Crossref] [PubMed]
- Dvir T, Timko BP, Kohane DS, et al. Nanotechnological strategies for engineering complex tissues. Nat Nanotechnol 2011;6:13. [Crossref] [PubMed]
- Lutolf MP, Gilbert PM, Blau HM. Designing materials to direct stem-cell fate. Nature 2009;462:433-41. [Crossref] [PubMed]
- Engelmayr GC, Cheng M, Bettinger CJ, et al. Accordion-like honeycombs for tissue engineering of cardiac anisotropy. Nat Mater 2008;7:1003-10. [Crossref] [PubMed]
- Eschenhagen T, Eder A, Vollert I, et al. Physiological aspects of cardiac tissue engineering. Am J Physiol Heart Circ Physiol 2012;303:H133-43. [Crossref] [PubMed]
- Tuan RS, Boland G, Tuli R. Adult mesenchymal stem cells and cell-based tissue engineering. Arthritis Res Ther 2003;5:32. [Crossref] [PubMed]
- Klebe RJ. Cytoscribing: a method for micropositioning cells and the construction of two-and three-dimensional synthetic tissues. Exp Cell Res 1988;179:362-73. [Crossref] [PubMed]
- Xu T, Kincaid H, Atala A, et al. High-throughput production of single-cell microparticles using an inkjet printing technology. J Manuf Sci Eng 2008;130:021017 [Crossref]
- D'Amore A, Yoshizumi T, Luketich SK, et al. Bi-layered polyurethane–extracellular matrix cardiac patch improves ischemic ventricular wall remodeling in a rat model. Biomaterials 2016;107:1-14. [Crossref] [PubMed]
- Okamoto T, Suzuki T, Yamamoto N. Microarray fabrication with covalent attachment of DNA using bubble jet technology. Nat Biotechnol 2000;18:438-41. [Crossref] [PubMed]
- Goldmann T, Gonzalez JS. DNA-printing: utilization of a standard inkjet printer for the transfer of nucleic acids to solid supports. J Biochem Biophys Methods 2000;42:105-10. [Crossref] [PubMed]
- Cui X, Breitenkamp K, Finn M, et al. Direct human cartilage repair using three-dimensional bioprinting technology. Tissue Eng Part A 2012;18:1304-12. [Crossref] [PubMed]
- Cui X, Breitenkamp K, Lotz M, et al. Synergistic action of fibroblast growth factor-2 and transforming growth factor-beta1 enhances bioprinted human neocartilage formation. Biotechnol Bioeng 2012;109:2357-68. [Crossref] [PubMed]
- Cui X, Gao G, Qiu Y. Accelerated myotube formation using bioprinting technology for biosensor applications. Biotechnol Lett 2013;35:315-21. [Crossref] [PubMed]
- Xu T, Jin J, Gregory C, et al. Inkjet printing of viable mammalian cells. Biomaterials 2005;26:93-9. [Crossref] [PubMed]
- Guillotin B, Souquet A, Catros S, et al. Laser assisted bioprinting of engineered tissue with high cell density and microscale organization. Biomaterials 2010;31:7250-6. [Crossref] [PubMed]
- Pepper ME, Seshadri V, Burg TC, et al. Characterizing the effects of cell settling on bioprinter output. Biofabrication 2012;4:011001 [Crossref] [PubMed]
- Pepper ME, Seshadri V, Burg T, et al. Cell settling effects on a thermal inkjet bioprinter. Conf Proc IEEE Eng Med Biol Soc 2011;2011:3609-12. [PubMed]
- Khalil S, Sun W. Biopolymer deposition for freeform fabrication of hydrogel tissue constructs. Mater Sci Eng C 2007;27:469-78. [Crossref]
- Murphy SV, Atala A. 3D bioprinting of tissues and organs. Nat Biotechnol 2014;32:773. [Crossref] [PubMed]
- Lantada AD, Morgado PL. Rapid prototyping for biomedical engineering: current capabilities and challenges. Annu Rev Biomed Eng 2012;14:73-96. [Crossref] [PubMed]
- Melchels FP, Feijen J, Grijpma DW. A review on stereolithography and its applications in biomedical engineering. Biomaterials 2010;31:6121-30. [Crossref] [PubMed]
- Chang CC, Boland ED, Williams SK, et al. Direct-write bioprinting three-dimensional biohybrid systems for future regenerative therapies. J Biomed Mater Res B Appl Biomater 2011;98:160-70. [Crossref] [PubMed]
- Miller JS, Stevens KR, Yang MT, et al. Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat Mater 2012;11:768-74. [Crossref] [PubMed]
- Lee VK, Kim DY, Ngo H, et al. Creating perfused functional vascular channels using 3D bio-printing technology. Biomaterials 2014;35:8092-102. [Crossref] [PubMed]
- Ott HC, Matthiesen TS, Goh SK, et al. Perfusion-decellularized matrix: using nature's platform to engineer a bioartificial heart. Nat Med 2008;14:213-21. [Crossref] [PubMed]
- Gu L, Feng J, Zhang D, et al. Bioengineering 3D Cardiac Microtissues Using Bioassembly. In: Serpooshan V, Wu S. editors. Cardiovascular Regenerative Medicine. Springer, 2019:107-23.
- Lu HF, Leong MF, Lim TC, et al. Engineering a functional three-dimensional human cardiac tissue model for drug toxicity screening. Biofabrication 2017;9:025011 [Crossref] [PubMed]
- Serpooshan V, Chen P, Wu H, et al. Bioacoustic-enabled patterning of human iPSC-derived cardiomyocytes into 3D cardiac tissue. Biomaterials 2017;131:47-57. [Crossref] [PubMed]
- Fang Y, Zhang T, Zhang L, et al. Biomimetic design and fabrication of scaffolds integrating oriented micro-pores with branched channel networks for myocardial tissue engineering. Biofabrication 2019;11:035004 [Crossref] [PubMed]
- Alvarez-Paino M, Amer MH, Nasir A, et al. Polymer Microparticles with Defined Surface Chemistry and Topography Mediate the Formation of Stem Cell Aggregates and Cardiomyocyte Function. ACS Appl Mater Interfaces 2019;11:34560-74. [Crossref] [PubMed]
- Hnatiuk A, Mercola M. Stars in the Night Sky: iPSC-Cardiomyocytes Return the Patient Context to Drug Screening. Cell Stem Cell 2019;24:506-7. [Crossref] [PubMed]
- Blinova K, Dang Q, Millard D, et al. International Multisite Study of Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes for Drug Proarrhythmic Potential Assessment. Cell Rep 2018;24:3582-92. [Crossref] [PubMed]
- Doss M. Cell-based regenerative therapy of heart failure: where do we stand. Cell Stem Cells Regen Med 2015; [Crossref]
- Rojas SV, Kensah G, Rotaermel A, et al. Transplantation of purified iPSC-derived cardiomyocytes in myocardial infarction. PLoS One 2017;12:e0173222 [Crossref] [PubMed]
- Mathur A, Loskill P, Shao K, et al. Human iPSC-based cardiac microphysiological system for drug screening applications. Sci Rep 2015;5:8883. [Crossref] [PubMed]
- Zhang L, Pan Y, Qin G, et al. Inhibition of stearoyl-coA desaturase selectively eliminates tumorigenic Nanog-positive cells: improving the safety of iPS cell transplantation to myocardium. Cell Cycle 2014;13:762-71. [Crossref] [PubMed]
- Rubach M, Adelmann R, Haustein M, et al. Mesenchymal Stem Cells and Their Conditioned Medium Improve Integration of Purified Induced Pluripotent Stem Cell–Derived Cardiomyocyte Clusters into Myocardial Tissue. Stem Cells Dev 2014;23:643-53. [Crossref] [PubMed]
- Kawamura M, Miyagawa S, Fukushima S, et al. Enhanced survival of transplanted human induced pluripotent stem cell–derived cardiomyocytes by the combination of cell sheets with the pedicled omental flap technique in a porcine heart. Circulation 2013;128:S87-94. [Crossref] [PubMed]
- Li H, Gao J, Shang Y, et al. Folic acid derived hydrogel enhances the survival and promotes therapeutic efficacy of iPS cells for acute myocardial infarction. ACS Appl Mater Interfaces 2018;10:24459-68. [Crossref] [PubMed]
- Masumoto H, Nakane T, Tinney JP, et al. The myocardial regenerative potential of three-dimensional engineered cardiac tissues composed of multiple human iPS cell-derived cardiovascular cell lineages. Sci Rep 2016;6:29933. [Crossref] [PubMed]
- Noor N, Shapira A, Edri R, et al. 3D printing of personalized thick and perfusable cardiac patches and hearts. Adv Sci (Weinh) 2019;6:1900344 [Crossref] [PubMed]
- El Harane N, Kervadec A, Bellamy V, et al. Acellular therapeutic approach for heart failure: in vitro production of extracellular vesicles from human cardiovascular progenitors. Eur Heart J 2018;39:1835-47. [Crossref] [PubMed]
- Liu B, Lee BW, Nakanishi K, et al. Cardiac recovery via extended cell-free delivery of extracellular vesicles secreted by cardiomyocytes derived from induced pluripotent stem cells. Nat Biomed Eng 2018;2:293-303. [Crossref] [PubMed]
- Heallen TR, Martin JF. Heart repair via cardiomyocyte-secreted vesicles. Nat Biomed Eng 2018;2:271-2. [Crossref] [PubMed]
- Sayed N, Liu C, Wu JC. Translation of human-induced pluripotent stem cells: from clinical trial in a dish to precision medicine. J Am Coll Cardiol 2016;67:2161-76. [Crossref] [PubMed]
- Laflamme MA, Gold J, Xu C, et al. Formation of human myocardium in the rat heart from human embryonic stem cells. Am J Pathol 2005;167:663-71. [Crossref] [PubMed]
- Caspi O, Huber I, Kehat I, et al. Transplantation of human embryonic stem cell-derived cardiomyocytes improves myocardial performance in infarcted rat hearts. J Am Coll Cardiol 2007;50:1884-93. [Crossref] [PubMed]
- van Laake LW, Passier R, Monshouwer-Kloots J, et al. Human embryonic stem cell-derived cardiomyocytes survive and mature in the mouse heart and transiently improve function after myocardial infarction. Stem Cell Res 2007;1:9-24. [Crossref] [PubMed]
- Fernandes S, Naumova A, Zhu W, et al. Human embryonic stem cell-derived cardiomyocytes engraft but do not alter cardiac remodeling after chronic infarction in rats. J Mol Cell Cardiol 2010;49:941-9. [Crossref] [PubMed]
- Chong JJ, Yang X, Don CW, et al. Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature 2014;510:273-7. [Crossref] [PubMed]
- Yamazaki D, Kitaguchi T, Ishimura M, et al. Proarrhythmia risk prediction using human induced pluripotent stem cell-derived cardiomyocytes. J Pharmacol Sci 2018;136:249-56. [Crossref] [PubMed]
- Fiedler LR, Chapman K, Xie M, et al. MAP4K4 inhibition promotes survival of human stem cell-derived cardiomyocytes and reduces infarct size in vivo. Cell stem cell 2020;26:458. [Crossref] [PubMed]
- Patel D, Stohlman J, Dang Q, et al. Assessment of proarrhythmic potential of drugs in optogenetically paced induced pluripotent stem cell-derived cardiomyocytes. Toxicol Sci 2019;170:167-79. [Crossref] [PubMed]
- Blinova K, Stohlman J, Vicente J, et al. Comprehensive translational assessment of human-induced pluripotent stem cell derived cardiomyocytes for evaluating drug-induced arrhythmias. Toxicol Sci 2017;155:234-47. [Crossref] [PubMed]
- Magdy T, Burmeister BT, Burridge PW. Validating the pharmacogenomics of chemotherapy-induced cardiotoxicity: What is missing? Pharmacol Ther 2016;168:113-25. [Crossref] [PubMed]
- Drawnel FM, Boccardo S, Prummer M, et al. Disease modeling and phenotypic drug screening for diabetic cardiomyopathy using human induced pluripotent stem cells. Cell Rep 2014;9:810-21. [Crossref] [PubMed]
- Burridge PW, Li YF, Matsa E, et al. Human induced pluripotent stem cell–derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nat Med 2016;22:547. [Crossref] [PubMed]
- Ronaldson-Bouchard K, Vunjak-Novakovic G. Organs-on-a-chip: a fast track for engineered human tissues in drug development. Cell Stem Cell 2018;22:310-24. [Crossref] [PubMed]
- Ribas J, Sadeghi H, Manbachi A, et al. Cardiovascular organ-on-a-chip platforms for drug discovery and development. Appl In Vitro Toxicol 2016;2:82-96. [Crossref] [PubMed]
- Weng KC, Kurokawa YK, Hajek BS, et al. Human Induced Pluripotent Stem-Cardiac-Endothelial-Tumor-on-a-Chip to Assess Anticancer Efficacy and Cardiotoxicity. Tissue Eng Part C Methods 2020;26:44-55. [Crossref] [PubMed]
- Lind JU, Busbee TA, Valentine AD, et al. Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing. Nat Mater 2017;16:303-8. [Crossref] [PubMed]
- Zhao Y, Rafatian N, Wang EY, et al. Engineering microenvironment for human cardiac tissue assembly in heart-on-a-chip platform. Matrix Biol 2020;85-86:189-204. [Crossref] [PubMed]
- Huebsch N, Loskill P, Deveshwar N, et al. Miniaturized iPS-Cell-Derived Cardiac Muscles for Physiologically Relevant Drug Response Analyses. Sci Rep 2016;6:24726. [Crossref] [PubMed]
- Zhang YS, Aleman J, Arneri A, et al. From cardiac tissue engineering to heart-on-a-chip: beating challenges. Biomed Mater 2015;10:034006 [Crossref] [PubMed]
- Bruyneel AA, McKeithan WL, Feyen DA, et al. Will iPSC-cardiomyocytes revolutionize the discovery of drugs for heart disease? Curr Opin Pharmacol 2018;42:55-61. [Crossref] [PubMed]
- Zhu WZ, Xie Y, Moyes KW, et al. Neuregulin/ErbB signaling regulates cardiac subtype specification in differentiating human embryonic stem cells. Circ Res 2010;107:776-86. [Crossref] [PubMed]
- Zhang Q, Jiang J, Han P, et al. Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals. Cell Res 2011;21:579-87. [Crossref] [PubMed]
- Devalla HD, Schwach V, Ford JW, et al. Atrial-like cardiomyocytes from human pluripotent stem cells are a robust preclinical model for assessing atrial-selective pharmacology. EMBO Mol Med 2015;7:394-410. [Crossref] [PubMed]
- Liu C, Oikonomopoulos A, Sayed N, et al. Modeling human diseases with induced pluripotent stem cells: from 2D to 3D and beyond. Development 2018;145:dev156166 [Crossref] [PubMed]
- Nair P, Prado M, Perea-Gil I, et al. Concise Review: Precision Matchmaking: Induced Pluripotent Stem Cells Meet Cardio-Oncology. Stem Cells Transl Med 2019;8:758-67. [PubMed]
- Matsa E, Burridge PW, Yu K-H, et al. Transcriptome profiling of patient-specific human iPSC-cardiomyocytes predicts individual drug safety and efficacy responses in vitro. Cell stem cell 2016;19:311-25. [Crossref] [PubMed]
- Chow M, Boheler KR, Li RA. Human pluripotent stem cell-derived cardiomyocytes for heart regeneration, drug discovery and disease modeling: from the genetic, epigenetic, and tissue modeling perspectives. Stem Cell Res Ther 2013;4:97. [Crossref] [PubMed]
- Kopljar I, Lu HR, Van Ammel K, et al. Development of a human iPSC cardiomyocyte-based scoring system for cardiac hazard identification in early drug safety de-risking. Stem Cell Reports 2018;11:1365-77. [Crossref] [PubMed]
- Yang X, Rodriguez M, Pabon L, et al. Tri-iodo-l-thyronine promotes the maturation of human cardiomyocytes-derived from induced pluripotent stem cells. J Mol Cell Cardiol 2014;72:296-304. [Crossref] [PubMed]
- Sun X, Nunes SS. Bioengineering Approaches to Mature Human Pluripotent Stem Cell-Derived Cardiomyocytes. Front Cell Dev Biol 2017;5:19. [Crossref] [PubMed]
- Tomlinson L, Lu ZQ, Bentley RA, et al. Attenuation of doxorubicin-induced cardiotoxicity in a human in vitro cardiac model by the induction of the NRF-2 pathway. Biomed Pharmacother 2019;112:108637 [Crossref] [PubMed]
- Stoppel WL, Kaplan DL, Black LD 3rd. Electrical and mechanical stimulation of cardiac cells and tissue constructs. Adv Drug Deliv Rev 2016;96:135-55. [Crossref] [PubMed]
- Ulmer BM, Stoehr A, Schulze ML, et al. Contractile work contributes to maturation of energy metabolism in hiPSC-derived cardiomyocytes. Stem Cell Reports 2018;10:834-47. [Crossref] [PubMed]
- Mummery CL. Perspectives on the use of human induced pluripotent stem cell-derived cardiomyocytes in biomedical research. Stem Cell Reports 2018;11:1306-11. [Crossref] [PubMed]
- Morizane A, Kikuchi T, Hayashi T, et al. MHC matching improves engraftment of iPSC-derived neurons in non-human primates. Nat Commun 2017;8:385. [Crossref] [PubMed]
Cite this article as: Huang Y, Wang T, López MEU, Hirano M, Hasan A, Shin SR. Recent advancements of human iPSC derived cardiomyocytes in drug screening and tissue regeneration. Microphysiol Syst 2020;4:2.


