
Flushing out the scattering: fast on-chip microfluidic clearing of microtissues
By generating three-dimensional microtissues or organoids that better mimic the in vivo cellular functions than the two-dimensional cell cultures, the organ-on-a-chip technologies have been playing an increasingly important role in fundamental biology and drug discovery (1). The fast-growing on-chip microfluidic field is accompanied by the high demand for on-site imaging of the microtissues’ morphology, function, and molecular signatures. Clearly, to image the three-dimensional microtissues, the imaging technique should ideally provide volumetric information, together with microscopic resolutions for cellular or even subcellular analysis (2). Although many imaging modalities possess three-dimensional imaging capability, only optical microscopy, primarily the confocal microscopy and multi-photon microscopy, has the required spatial resolution and molecular/chemical sensitivity, and thus is widely used with on-chip microfluidic systems (3). However, the biological tissues are notoriously unfriendly to photons, with strong optical scattering resulted from the inhomogeneous distribution of subcellular structures, including cell membranes, nuclei, and mitochondria (4). High-resolution optical microscopy often suffers from the strong optical scattering of tissues, leading to a shallow imaging depth of a few hundred micrometers.
So far, there exist roughly two groups of methods to overcome the penetration limitation of optical microscopy: one involves improving the imaging system by using longer light wavelengths (5), designing more efficient optical detection (6,7), or correcting the phase of the incidental light waves (8); the other is to clear the tissue by physically or chemically reducing the optical scattering, i.e., making the tissue transparent (9,10). Compared with the efforts of improving the optical imaging systems, the optical clearing methods can better solve the imaging depth issue and can be readily implemented with traditional optical microscopy. On the other hand, the optical clearing methods usually suffer from strong cytotoxicity and thus cannot be applied for live cell imaging, which, however, is less a problem if only the morphologic information is of interest. Nevertheless, based on passive mass diffusion of the clearing agents into the tissues, optical clearing is still not widely adapted for three-dimensional optical imaging in on-chip microfluidic systems, due to the extremely long clearing time involved in the multi-step process. For example, it takes days even for the most advanced CLARITY technology to clear up a small piece of mouse brain (11). With such a low throughput, it is very challenging to apply the optical clearing technology on microfluidic platforms, which usually involves a closed system and in many cases presence of multiple microtissues. Therefore, there is an urgent demand for an innovative optical clearing method that suites the high-throughput imaging need of on-chip microfluidic systems.
Chen and the colleagues have recently developed a novel technology that has substantially accelerated the speed of optical clearing and simplified the process for the on-chip microfluidic system (12). This idea exploits the well-known physical phenomenon that the mass diffusion rate can be increased by an external fluidic pressure. The same physical mechanism also leads to the fact that a fast-blowing wind can quickly dry up a water spill on the road than that in a windless day. As shown in Figure 1A, Chen’s method can be implemented on an on-chip microfluidic system in three steps: first, a monomer solution is infused into the device, which preserves and fixes the tissue after gelation; second, a solution containing sodium dodecyl sulfate (SDS) is infused to wash away the cellular membranes of the tissue, exposing the intracellular proteins for fluorescent labeling; finally, a refractive index-matching solution (RIMS) is infused to eliminate the scattering interfaces within the tissue, allowing the tissue core to be visualized and imaged. Because of the active perfusion exerted by the microfluidic system, the authors have demonstrated an enhanced exchange rate of interstitial fluids by 567-fold, leading to an eventual 20 times faster clearing rate in three-dimensional microtissues. Such an impressive clearing speed has allowed the authors to image the segregation and compartmentalization of different cells during the formation of tumor spheroids (Figure 1B), and to track the degradation of vasculature over time within extracted murine pancreatic islets in static culture.
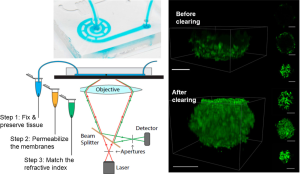
Compared with previous optical clearing methods (10), the major advantages of the microfluidic tissue clearing are 2-fold: (I) the same microfluidic system equipped with traditional optical microscopy can be used for both microtissue culturing and clearing, which has substantially simplified the clearing and imaging process, and (II) the microfluidic-enhanced mass diffusion significantly accelerates the clearing rate, allowing high-throughput study of the entire microtissues. This new method, demonstrated by several proof-of-concept applications, has opened the door for numerous assays that can tremendously benefit from high-throughput, on-chip, deep-penetration screening of three-dimensional microtissues, including but not limited to drug screening for multi-cellular biological functions and precision medicine that explores the heterogeneous tissue responses.
Acknowledgments
Funding: J Yao acknowledges the support of Duke MEDx grant.
Footnote
Provenance and Peer Review: This article was a standard submission to the journal. The article did not undergo external peer review.
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/mps.2017.10.01). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhang YS, Khademhosseini A. Seeking the right context for evaluating nanomedicine: from tissue models in petri dishes to microfluidic organs-on-a-chip. Nanomedicine (Lond) 2015;10:685-8. [Crossref] [PubMed]
- Smith LE, Smallwood R, Macneil S. A comparison of imaging methodologies for 3D tissue engineering. Microsc Res Tech 2010;73:1123-33. [Crossref] [PubMed]
- Pampaloni F, Ansari N, Stelzer EH. High-resolution deep imaging of live cellular spheroids with light-sheet-based fluorescence microscopy. Cell Tissue Res 2013;352:161-77. [Crossref] [PubMed]
- Wang LV, Wu HI. Biomedical Optics: Principles and Imaging. Hoboken, N.J.: Wiley-InterScience, 2007.
- Hong G, Lee JC, Robinson JT, et al. Multifunctional in vivo vascular imaging using near-infrared II fluorescence. Nat Med 2012;18:1841-6. [Crossref] [PubMed]
- Theer P, Denk W. On the fundamental imaging-depth limit in two-photon microscopy. J Opt Soc Am A Opt Image Sci Vis 2006;23:3139-49. [Crossref] [PubMed]
- Horton NG, Wang K, Kobat D, et al. In vivo three-photon microscopy of subcortical structures within an intact mouse brain. Nat Photonics 2013;7:205-9. [Crossref] [PubMed]
- Horstmeyer R, Ruan HW, Yang CH. Guidestar-assisted wavefront-shaping methods for focusing light into biological tissue. Nat Photonics 2015;9:563-71. [Crossref] [PubMed]
- Ke MT, Fujimoto S, Imai T. SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci 2013;16:1154-61. [Crossref] [PubMed]
- Zhu D, Larin KV, Luo Q, et al. Recent progress in tissue optical clearing. Laser Photon Rev 2013;7:732-57. [Crossref] [PubMed]
- Chung K, Deisseroth K. CLARITY for mapping the nervous system. Nat Methods 2013;10:508-13. [Crossref] [PubMed]
- Chen YY, Silva PN, Syed AM, et al. Clarifying intact 3D tissues on a microfluidic chip for high-throughput structural analysis. Proc Natl Acad Sci U S A 2016;113:14915-20. [Crossref] [PubMed]
Cite this article as: Yao J. Flushing out the scattering: fast on-chip microfluidic clearing of microtissues. Ann Clin Oncol 2017;1:3.


