Organs-on-chip monitoring: sensors and other strategies
Introduction
Throughout the ages, our curiosity for decoding the secrets of human body have skyrocketed which in turn led to many advancements in various scientific disciplines. In that regard, one of the fields that witnessed a paradigm shift is tissue engineering. From innovative methods of genetic manipulation via clustered regularly interspaced short palindromic repeats (CRISPR) technology to the investigation of advanced biomaterials to improve the efficacy of the scaffolding biomaterials tremendous progress have been achieved (1). As one of the advancements that came into being by 2010, “organs-on-chip” (OOC) are considered as very promising candidates for investigating mechanism of organ physiology as well as drug testing platforms with future advancements to be used for personalized medicine and disease modelling (Figure 1).
The concept of OOC was introduced by Donald Ingber (2). The term OOC is used to define a microfluidic culture that contains continuously perfused chambers inhabited by living cells to simulate the activities, mechanics and physiological response of tissue, organs and organ systems (3). Dr. Ingber and his team at Wyss Institute introduced the first model for “lung-on-a-chip”, a microfluidic chip that represents a breathing and immune-reactive lung composed of human airway, capillary and immune cells (2). Following this work, other early examples of OOC, “intestine-on-a-chip” (4), “lymphe node-on-a-chip” (5), blood vessels-on-a-chip (6) have been launched as can be seen in Figure 2. And followed by more complicated platforms that connects two or more organs (7-9).

As the complexity of OOC systems increases to be able to create pharmacologically and physiologically relevant models, the necessity to integrate relevant assessment methods into such platforms also increases. In order to be able to provide spatially and temporally resolved information about cell physiology and microenvironment as well as pharmacodynamics drug responses, sophisticated monitoring tools and read out of in vitro systems are required (10). In that regard, engineering approaches have been used to develop physical, chemical and biological sensors can be integrated to OOC. These sensors have been shown to provide reproducible results in a short time with data transmission, multiplexing and on-line monitoring capability by analysing very low volume of samples (9,10). Readout technologies that has been suggested and used so far for OOC platforms are based on measurement of physical parameters associated with tissue/organoid microenvironment (such as O2, pH, CO2 and osmolarity), biological properties (protein and metabolite secretion, DNA methylation etc.), morphology (cell layer barrier, cell-cell interaction, via fluorescence and confocal microscopy).
The purpose of this review paper is to provide a critical and comprehensive report on sensors integration on OOC platforms. After the section as a mini-review on current OOC technologies, several sensor designs have been introduced and detailed by figures.
State-of-the-art OOC
The majority of scientists and researchers usually rely on biological hypotheses on primary cultures and in vitro cell culture platforms to investigate biological processes and develop therapeutic strategies, focusing their attention to the biological response of the cells, without tight control of the biological context surrounding them, but this simplicity means that they fail to mimic key aspects of the human body (11). In contrast, industry usually test drugs in vivo, but a high percentage of principal compounds fail during the clinical trial phase because of their inherently low-throughput (12-14). Although these approaches have been successful, they have significant shortcomings. Despite the fact that in vivo models can produce integrated multi-organ responses which are impossible to achieve using conventional in vitro models, isolating the prominent tissues or cell groups corresponding to a particular physiological or pathophysiological response is difficult and leads to further complications and shortcomings. In addition to the ethics surrounding in vivo model usage, serious concerns exist over their biological relevance to humans (13). On the other side, in vitro platforms cannot often simulate the complex cell-cell and cell-matrix interactions crucial for regulating cell behaviour in vivo, they are useful for studying the molecular basis of physiological and pathological responses. Because of these limitations, a significant number of new drug candidates fail to make into the market due to low efficiency or severe side effects. These issues together with regulatory restrictions that limits the use of animal models, have generated considerable interest in improving human-based tissue-like constructs for disease modelling and drug and chemical testing, that will soon revolutionize the way researchers perform their investigations and pharmaceutical companies test their drugs (15,16). A requirement for industrial use of new three-dimensional (3D) in vitro models which are closely mimicking human normal and pathological organs and tissues is scalability and also may include cell and tissue geometries, electrical activity and substrate mechanics. OOCs are models that can provide these new models by combining sophisticated chip technology with biology. This can modulate human responses to be recapitulated in vitro more accurately than in other systems described so far (11). Not only can OOC devices provide the biological relevance but they are also the requisite of high throughput applications because of their small scale, reliability of results and low costs and their ability to provide new tools for controlling the transport and availability of chemical and biochemical signals on micro-scales (17). Microfabrication approaches such as photolithography, soft lithography, microcontact printing and micromolding are enabling more-complex tissue cultures to be patterned on-chip (18-20).
Microfluidic systems offers unprecedented and enhanced dynamic control over culture conditions and the cellular microenvironment within on-chip tissue models, and this technology is currently in a mature state to provide nutrients and dissolved gasses and to apply stimuli such as chemical gradients, spatial homogeneity, time-dependent biochemical stimulations, and substrate mechanical properties (18). The resulting independent and functional tissues shaped product with a morphology that attempts to recapitulate in vivo organs conditions has led to the concept of ‘OOC’ (17). Incorporating stimulation and sensing technologies on-chip and interfacing these with cultured cells is another important step for the real-time manipulation and detection of cellular behaviour (18). Miniaturization to the micrometer scale offers several advantages such as superior control over the local cellular microenvironment and significant reduction in the use of compounds, reagents and cells, therefore increasing the experimental throughput in a cost effective manner (21).
By developing the single-OOC systems for long-term ex vivo culture, the significance of interactions between different cell types and cell-ECM interactions within the same functional unit of the organ, as well as stimuli of the microenvironment surrounding the extracted tissue in vivo (in order to maintain its functionality in vitro) were investigated (14). As biosensors can sense specific biological molecules within the miniaturized tissue constructs in real-time, at very low concentration levels, through ultrasensitive optical, electrochemical, or acoustic sensing systems, they are gradually becoming an integral part of such tissue engineering systems particularly in microfluidic tissue engineering models (22).
Liver-on-a-chip
The major reason of drug withdrawal is the drug-induced hepatotoxicity and hence there is a huge demand for development of a robust in vitro model for testing hepatotoxicity as well as assessment of drug metabolism. Thanks to recent advancements in microfabrication techniques as well as 3D bioprinting (23,24), various liver-on-a-chip platforms have been developed so far (25-29). These platforms are made from optically transparent chips with channels (50–500 µm wide) where various liver cell types such as hepatocytes, non-parenchymal cells or other stromal cells have been cultured in either 2D or 3D format as well as mono or co-cultures (30) .
Brain-on-a-chip
As being one of the most complex organs, brain has been of great interest for decades to medical scientists whose research interests are on neuroscience, neurodegenerative diseases, electrophysiological, and pharmacological studies on blood-brain barrier. Thus, various attempts have been put to develop in vitro platforms to investigate human neuronal differentiation and chemotoxicity (31), modeling Alzheimer’s disease (32), traumatic brain injury (33) and blood-brain barrier (34).
Lung-on-chip
One of the main causes of death is lung disease, and the rate of pulmonary diseases has been increasing for decades. The only treatment for these types of diseases is lung transplantation. However, the lack of donors is the main limitation of this method. Therefore, investigating the new strategies with the aid of tissue engineering and microfluidics techniques for functional analysis and drug screening is needed (35). Mimicking the functional unit of the lung, alveolar-capillary interface, is challenging because of the difficulties of replicating the structural and functional properties of the system while simulating the mechanical changes associated with normal breathing. Huh et al. designed a flexible porous membrane and cultured human alveolar epithelial cells on one side and human pulmonary microvascular endothelial cells on the other. To mimic the dynamic mechanical stimulation of the cells caused by breathing, the membrane stretched using a vacuum to neighbouring chambers. The device has the potential for drug screening for lung disorders and also investigating the impact of toxins (2).
Heart-on-a-chip
Heart disease causes 1 out of 4 of all deaths in the United States. So, developing the cardiac drug, side effects of drugs and the interactions between multiple drugs has been a strong emphasis on the medical community. Microscale on-chip solutions offer an attractive platform to conduct cost-efficient and time-efficient drug assays before the clinical trials. Some examples of such a microscale solution have been provided by some researches. For example at Parker’s work, heart muscle cells were cultured onto small tissue strips to create a functional engineered beating cardiac tissue on a flexible substrate using microfabrication technology. Then these structures were exposed to drugs and electrically stimulated. So, researchers were enabled to evaluate the cardiotoxic effects of the drugs (36). Aubin et al. have encapsulated cardiomyocytes inside hydrogels with different micropatterns to create the 3D aligned fiber-like structures of cardiac tissues for drug testing (37). In another study, embryonic cardiac cells were cultured inside a bioreactor and exposed to pulsatile flow and varying strains to mimic the cardiac cycle in the left heart ventricle to conducting drug tests (38). Personalized medicine could be achievable using on-chip systems for drug testing on patients’ own cells to maximize the success of treatment (39). Using bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium is another attempt to create an in vitro hear-on-a-chip model for drug screening and disease modelling (40). In another study, a heart-on-a-chip platform was developed, which recapitulates the physiologic mechanical environment experienced by cells in the native myocardium. The device includes an array of hanging posts to confine cell-laden gels, and a pneumatic actuation system to induce homogeneous uniaxial cyclic strains to the 3D cell constructs during culture to generate mature and highly functional micro engineered cardiac tissue (41).
Bladder-on-a-chip
Bladder cancer is the most frequent cancer of the urinary system and was found to be caused by many factors. Therefore, finding a way to diagnose this cancer at early stage is really critical to increase the survival rate of the patients. To treat the bladder cancer, currently, a method of incising part of the body is being used, but it has difficulty in diagnosing bladder cancer at an early stage. (https://www.google.com/patents/US9359646). A research group has worked on the quantitative detection of Galectin-1 (Gal-1) protein, a biomarker for the onset of multiple oncological conditions, especially bladder cancer. An ultrasensitive and real-time impedance based immunosensor has been fabricated which consists of a gold annular interdigitated microelectrode array patterned, with the ability to electrically address each electrode individually. To improve sensitivity and immobilization efficiency, trapped nanoprobes (Gal-1 antibodies conjugated to alumina nanoparticles through silane modification) on the microelectrode surface were used. The normalized impedance variations reveal a linear dependence on the concentration of cell lysate present while specificity is demonstrated by comparing the immunosensor response for two different grades of bladder cancer cell lysates. Also, they designed a portable impedance-analyzing device to connect the immunosensor for regular check-up in point of care testing with the ability to transfer data over the internet using a personal computer.
Uterus-on-a-chip
The female reproductive tract is a complex system consisting of ovaries, fallopian tubes, uterus and cervix. All these major organs participate in an autonomous or interdependent way for the production of ova, secretion of sex hormones and the maintenance of pregnancy throughout the gestation (42). The reproduction of a total uterine system on a microfluidic chip might be of great impact for in vitro fertilization-embryo transplantation (IVF-ET) (43) and for testing the efficacy and toxicity of new drugs (42). After various attempts to mimic the physiological function of the reproductive system (43,44) recently an advanced and extremely attractive and promising system has been realized by Xiao et al. (42), the so called EVATAR (“avatar”, digital representation of an individual, plus the name “Eve”). This powerful tool phenocopies the human menstrual cycle and pregnancy by reproducing the endocrine loops between organ modules for the ovary, fallopian tube, uterus, cervix and liver, with a sustained circulating flow between all tissues.
Vessels-on-a-chip
The blood vascular system consists of blood vessels (arteries, arterioles, capillaries, and veins) that convey blood from one organ to another ensuring a correct the organ’s functionality. Therefore, blood vessels represent the basic and vital building blocks of this system (45) since they deliver nutrients and oxygen to tissues, and remove waste products too. To correctly mimic the vascular physiology, morphogenesis and development mechanical stimulation is an indispensable factor therefore it is needed to provide microfluidic chip capable of delivering fluid shear stress (FSS) and cyclic stretch (CS) simultaneously or independently (46). Moreover, vessels-on-a-chip reconstruction is not only useful to connect each-others the various compartments of a body-on-a-chip application (45), but also to study system dysfunctions caused by specific disease, e.g., angiogenesis in patients with tumors, diabetes or with wounds (47), or to study the effects of inflammation on vascular integrity or interrupt flow, e.g., thrombosis (48).
Needs for sensors in OOCs
Although the field of OOCs evolves over time, there is still need for more sophisticated, sensors integrated OOCs that will provide continuous information about the viability and metabolic activity of the tissue constructs/organoids real time. To achieve such a goal, several criteria should be ensured. One of the most important criteria is to integrate sensors to monitor physical and biochemical parameters associated with functionality of OOC models. In this section, the needs of sensors for OOC platforms will be discussed in detail to give insights about the next section where the current monitoring strategies were explained.
Sensors to monitor culture environment
The major determinant for the formation of functional 3D organoid/tissue is cell culture environment with all its parameters such as temperature, pH, humidity and oxygen level and nutrient content. In their microenvironment, cells are exposed to changes of these parameters as well as gradients of secreted metabolites from neighbouring cells, and mechanical interactions from cell-cell and cell-extracellular matrix (ECM) contact. Integration of analytical devices to 3D cell culture platforms as well as OOCs can enable controllable and reproducible culture environment.
Sensors to monitor cell behavior
Monitoring cell behaviour is as equally important as cell culture media monitoring since cell isolation and proliferation are critical steps for building up a 3D tissue construct. Not only identifying the cell density and viability but also investigation of cellular function by cultivation time and upon any stimulus are needed. So far, sensors for the purpose of monitoring cellular adhesion (49), detachment (50), death (51), response to osmotic stress (52), sepsis (53) have been developed based on widely used surface plasmon resonance (SPR) technique, photonic crystal fibers (54), and resonant waveguide grating EPIC® systems (55). Contractile properties of cells, especially cardiomyocytes for heart-on-a-chip platforms is of great importance and it is reported to be affected by intracellular calcium secretion. Hence, it is important to measure the calcium levels for heart-on-a-chip platforms (56,57).
Sensors to monitor stimulations
Well representation of mechanical, chemical, electrical and other interactions that affects the functions of human organs haven’t been achieved yet due to inherent spatial and temporal features of human body. In order to provide physiologically relevant organ mimicry, OOC platforms are required to integrate all types of stimulation that any organ is exposed to in vivo. Besides accurate replication, monitoring of these mechanical cues in vitro is also an urgent need for OOC platforms.
Sensors to monitor mechanical stimulations
Time-varying mechanical deformation is of great importance in respiratory and circulatory systems since air and blood are pumped by lung and heart. To simulate the cyclic expansion and contraction in the lung, Huh et al. developed a lung-on-a-chip platform where a vacuum pump is used to achieve cyclic stretching and relaxation of the elastic membrane and device wall of PDMS (2). Similar technology has also been used for the design of gut-on-a-chip to mimic the trickling fluid flow as well as cyclic peristaltic motions that a gut experiences in vivo (4). Creation of actively deformable membranes is needed for vasculature mimicries due to pulsatile motion of blood and cyclic stretching. It is shown that, by a mechanical stretch applied via microfluidic chip where a deformable membrane on which cells are cultured can modulate stem cell differentiation to vascular smooth muscle cells (58).
Sensors to monitor chemical gradients
Chemical stimulation is another parameter that should have taken into account to develop and maintain physiologically relevant OOC platforms. Thanks to microfluidic devices, it is easy to control the fluid flow to tune chemical flux at the same scale as in the case of in vivo. When these microfluidic devices are integrated with detection platforms, it will be possible to enlighten the function of chemical factors over cell-cell interactions (59). Creating a well representative of a tissue microenvironment is highly dependent on spatiotemporal control of chemical factors such as growth hormones and cytokines. Such a control has been achieved via a microfluidic device that varies the extracellular environment through the measurement and feedback control of pressure in the fluid pumping mechanism (60). It is also shown that, by using a small amount of chemicals, it is possible to create, monitor and control an oxygen gradient inside microfluidic channels for cell culture (61).
Sensors to monitor electrical stimulations
Electrical and electromechanical characterization and control of cellular microenvironment is becoming increasingly important in recent years especially for central nervous system (CNS) and muscular system such as heart. In the early developmental phase, the brain receives large amounts of electrical stimulation critical for developing excitatory synapses. In addition to that, functional electrical stimulation is proved to promotes regeneration of spinal cord injury (62). In another study, researchers showed that by applying electrical stimulation via platinum electrodes, it is possible to achieve myelinsegment formation (63). When it comes to cardiac tissue engineering and heart-on-a-a-chip platforms, electrical stimulation is reported to be crucial since it stimulates the mechanical cycling of heart and when applied in microfluidic devices, results in cellular reorganization and transformation (64). Therefore, researchers have developed specialized devices to recapitulate the unique microenvironment of heart in vitro (65). Moreover, stem cell differentiation into cardiac tissue has also been reported in many papers (65,66). However, once a heart-on-a-chip platform with electrical stimulation has been engineered, in vitro assessment of the efficacy is required, so as the sensor integration for measurement of electrical signal as well as cell contractility.
Wireless and portable monitoring feature towards IoT systems
The possibility to enable real-time, quantitative information of the cell status is one of the most important aspects for monitoring the cellular processes. Indeed, by providing a wireless and portable system able to continuously monitor the cell cultivations, it would be possible to constantly control the cultivation parameters enabling a prompt modification in case of necessity (67). To enable this remote on-line monitoring, portable devices have been adopted. In particular, dedicated application has been developed to enable data-visualization (68) and set-up hardware control (69) from tablet or smart-watch. Most recently, also wearable technology was introduced in medical and biomedical applications. In particular, Google Glass (70) has been adopted to display information in a smartphone-like hands-free format exploiting voice recognition commands.
Another possibility is to wear smart-watches connected with the sensors integrated to the organ-on chip device realizing an IoT monitoring system. In this way, monitoring parameters reachable from any place and at any time just wearing the smart-device. Moreover, alerts can be programmed and send to the user in order him to intervene the ongoing experiment (71).
In terms of OOC platforms, IoT systems could be useful in future where long term monitoring such as pH, morphological changes are required.
State-of-the art: current biosensors for OOCs
Although the need for sensors integration ot monitor OOCs is urgent, there is not much in literature that exemplify real integration of OOCs with sensors. Hence, in this review, we also included sensors designed for cell culture monitoring considering their potential to be integrated to OOC platforms. Classification of sensors was made according to the purpose of monitoring such as detection of metabolism products, monitoring of physical parameters and cell fate.
Sensors for detection of metabolism products
Cellular metabolic activity is the combination of catabolic and anabolic pathways and sensors in this context is required to assess the efficiency of metabolic activity by providing information about concentration of certain products. For instance, extracellular oxygen, carbon dioxide, glucose and lactate are needed for monitoring and understanding how well the cell is carrying out respiration. Similarly, hydrogen peroxide (H2O2) has been reported to have an important role in normal cellular growth and proliferation. Toxicity is another concern when it comes to drug testing and should be tested upon monitoring cell type-specific molecular biomarkers such as albumin and transferrin in the case of hepatotoxicity (72) and creatine kinase-MB for cardiac toxicity (73).
Hydrogen-peroxide sensors
As being one of the reactive oxygen species (ROS), H2O2 plays an important role in cellular processes (74) especially in proliferation since its long lifetime allows cell penetration that result in series of cell damage (75). Hence, selective and sensitive detection of H2O2 both inside and outside of the cells are essential to discover the affects in cellular level as well as diagnosis of real time conditions of the cells. Fast and reliable monitoring of H2O2 is of great interest to many fields including pharmaceutical and clinical and therefore a variety of techniques have been developed for the monitoring of H2O2 such as colorimetry (76,77), chemiluminescence (78), fluorescence (79-81), electrochemical methods (82,83). Among them, fluorescence and electrochemical methods have received a considerable interest. More than that, when the H2O2 is detected with electrochemical sensors, it opens the way to also detect a series of other cellular metabolites that are usually metabolised by oxidases.
Fluorescence based H2O2 detection
The beauty of using fluorescence probes for H2O2 monitoring is to be able to do detection in live cells, tissues and even animals. Various fluorescent based H2O2 sensors have been developed so far (79-81,84). The rationale of sensing is either based on the usage of proteins that has intrinsic fluorescence signal that is changed due to exposure of ROS like H2O2 or to the usage of metal nanoclusters such as gold nanoclusters (AuNCs) or silver nanoclusters (AgNCs). These NCs are modified with enzymes to trigger catalysis function for certain analytes such as in the case of horseradish peroxidase (HRP) to be quenched in the case of H2O2. For instance, usage of NCs on fluorescent based sensing was demonstrated by a group who designed denatured lysozyme capped silver nanoclusters (dLys-AgNCs) where lysozyme had a role of stabilization as well as signaling element towards OH. Hence, this sensor was used for detection of H2O2 in live cells to monitor oxidative dama to proteins (84) (Figure 3A).
In another work, Zhang et al. have synthesized HRP-AuNCs (Figure 3B) and achieved fluorescence detection of H2O2 over the range of 100 nM–100 µM (81). After that, Yang Tian and his groups managed to quantify H2O2 in living cells by using different strategies. In one of the studies, they designed AuNCs protected by a bovine serum albumin (BSA) which is employed as a reference fluorophore and the organic molecule 2-[6-(4'-hydroxy)phenoxy-3H-xanthen-3-on-9-yl] benzoic acid which acted as recognition element that gives emission after interaction with •OH. The sensor that has the ~0.68 µM detection limit was applied to monitor oxidative stress in live cells (80) (Figure 3C). The same group designed another ratiometric fluorescent biosensor for detection of ROS species, namely O2•–, this time by using carbon dots as the fluorophore and hydroethidine as recognition molecule. The sensor showed a lower detection limit (100 nM) than the previous one and again used for monitoring oxidative stress for HeLa cells (Figure 3D) (79).
Electrochemical detection
Electrochemical techniques have taken great interest for H2O2 detection mainly because they require simple instrumentation and provide sensitive and selective detection in a short assay time. Most of the electrochemical H2O2 biosensors were based on catalytic activity of HRP but due to its serious disadvantages over practical applications, non-enzymatic sensors have been developed (85) by employing metal nanoparticles (86), carbon based nanomaterials (graphene, carbon nanotubes etc.) (87). So far, considering cell-monitoring applications, electrochemical H2O2 biosensors for the purpose of detection extracellular H2O2 released from human breast cancer cells MCF-7 (88), HeLa and HepG2 cells (89), Raw 264.7 cells (82), PC12 cells (83).
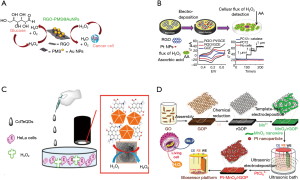
Even if metal nanoparticles such as gold nanoparticles (AuNps) provide good electrical signal, most of the times developed sensors does not meet with the requirement of sensitivity for cell based applications which needs nanomolar concentration range detection of H2O2. Hence, researchers are directed to develop nanocomposites with various nanomaterials. For instance, a hybrid nanomaterial was fabricated by immobilization of AuNpS onto mesoporous silica (PMS) coated reduced graphene oxide (RGO) (Figure 4A) to be able to measure H2O2 secretion by HeLa and HepG2 cells. The sensor showed a low detection limit of 60 nM with a good selectivity (89). Similarly, other group have again used RGO to create RGO-Pt nanocomposites on glassy carbon electrode (Figure 4B) for PC12 (rat adrenal medulla pheochromocytoma) cells H2O2 secretion monitoring (Figure 4C) (90). Another example of Pt nanoparticle to make hybrid with carbon nanomaterial was presented by the work of Liu et al. (92) who used Pt nanoparticles (PtNPs) decorated porous graphene (PG) to monitor HeLa cells up to 0.50 µM of H2O2 secretion. Although these biosensors offer good detection limit and has cell monitoring capability, making them compact to be integrated to OOC platforms is tricky. Flexible biosensors or fluidic-based biosensors based on polymer substrates such as PDMS could be of great interest in that regard. For instance, Duan et al. developed a flexible electrochemical H2O2 sensor by fabricating Pt nanoparticles on freestanding reduced graphene oxide paper (rGOP) carrying MnO2 nanowire networks (Figure 4D) (91). The sensor reported to have 1.0 µM detection limit and used for monitoring oxidative damage to human liver cancer cells. Another interesting work done via graphene shows a simple strategy for non-enzymatic H2O2 detection from human serum samples and that released from human cervical cancer cells (Figure 4C). The sensor is developed by AuNPs on graphene quantum dots (AuNPs-N-GQDs) and showed detection limit of 0.12 µM (90).
Glucose and lactate sensors
Glucose, the primary energy source of cells, is metabolized first into pyruvate, an intermediate compound that will further be converted to acetyl-CoA and fed into the citric acid cycle under aerobic conditions. However, under anaerobic conditions, or due to shortage of oxygen, accumulated pyruvate is reduced to lactate. For example, in myocardial ischemia or congestive heart failure, the lactate secretion is increased. Hence, increase in lactate concentrations is considered as an indication of disruption of citric acid cycle mainly because of oxygen shortage. Due to the fact that metabolic toxins as well as chemical agents and drugs affects enzymatic reactions occur during citric acid cycle, it is very important to monitor glucose and lactate concentration for drug toxicity studies. It is also very essential to have glucose and lactate sensors for any OOC platforms that aims to perform toxicity studies.
In addition to that, cancerous cells are known to have a metabolism change as a result of up-regulated glycolysis because of mitochondrial activity suppression. This suppression cause increase in glucose uptake and lactate production despite sufficient oxygenated conditions. This phenomenon that is known as aerobic “glycolysis” or “warburg effect” affects not only intracellular but also extracellular environment since lactate is exported to the cytoplasm and lowers the pH (93). Moreover, studies have shown that, increased intratumoral lactate concentration result in metastasis in many cancer types (93). Therefore, lactate is considered as an important biomarker for cancer malignancy.
Up to now, various types of glucose and lactate sensors, mainly optical (94-97) and electrochemical (97-102) have been developed to provide information about cell viability upon toxicity (97,98,103,104), cancer cell progression, deciphering cancer metabolic pathways (94), better understanding of molecular basis of brain functions (99) or different metabolic models of hearts including those of hypoxia and ischemia (100), measuring (cancer) cell metabolism (95,96,101,102) and tracking the dynamics of mitochondrial dysfunction (97).
Electrochemical glucose and lactate sensors
Electrochemical glucose and lactate biosensors incorporate enzymes such as glucose oxidase (GOX) or lactate oxidase (LOX), lactate dehydrogenase (LDH) as bio-recognition element to metabolize the corresponding target (eqn.1 and 2) and produce H2O2 of which electrochemical signal is measured by time via voltammetric or amperometric methods.
Electrochemical sensors gained considerable interests in recent years due to their microfabrication enable ease of integration to microfluidic devices. In addition to that, multiplexed sensors made it possible to do multiparametric monitoring of metabolites. One example to such multiplexed microfluidic based electrochemical sensors is presented by Mao et al. (Figure 5A) who managed continuous and simultaneous monitoring of glucose, lactate and ascorbate in the rat brain to monitor brain ischemia (99). Toxicity monitoring caused by drugs is another aim for development of EC biosensors. In that regard, a toxicity monitoring study aimed to observe changes in hepatic tumor cell upon exposure to mitochondrial inhibitor rotenone via enzyme-based electrochemical flat-plate sensors. The system (Figure 5B) was reported to provide real-time information on drug induced liver toxicity in vitro over several days (104).
A recent work by Weltin et al. shows the integration of EC microsensor system into a 3D human HepaRG hepatocyte spheroids for continuous and long-term monitoring of lactate production and oxygen consumption upon exposure to a drug Bosentan, that is used to treat pulmonary hypertension (103).
Monitoring cancer cell metabolism is another aim for EC glucose and lactate biosensors. Weltin et al. developed multiparametric microphysiometry system (Figure 5C) for monitoring the metabolism of T98G human brain cancer cells that are cultivated on chip. In addition to measuring glucose consumption and lactate production, system also allowed measurement of pH and oxygen in the cell culture microenvironment (102). Likewise, researchers developed enzyme-based lactate biosensors into a microfluidic network of hanging drops of human colon carcinoma spheroids for continuous in-situ monitoring of several effects such as culture conditions or dosage of compounds (105).
Optical glucose and lactate sensors
Optical glucose and lactate sensors have advantages over electrochemical biosensors due to their label-free, real-time and continuous and long-term monitoring schemes. For development of such sensors, fluorescent dyes, quantum dots, crystalline colloidal arrays, plasmonic nanoantennas are incorporated (106).
Cell monitoring applications of optical glucose and lactate sensors are mainly focused on cancer cell biology. Especially, for studying tumour progression, platforms that allow monitoring in single cell level is of great importance. Li et al. a nanoscale optical fiber lactate was developed for monitoring extracellular lactate concentrations of single HeLa, MCF-7, and human fetal osteoblast (hFOB) cells. Various lactate inhibitors were tested via sensors to show the potential of the system for evaluation of metabolic agents on cancer survival (96). Recently, Abbyad et al. developed present a droplet microfluidic method that allows monitoring in K562 leukemia and U87 glioblastoma cancer cell lines. Group also examined the lactate efflux under the chemical inhibition via α-cyano-4-hydroxycinnamic acid (94).
Biosensors for cell secreted molecules detection
Monitoring OOC platforms is not only covers monitoring of physical parameters but also biochemical ones. In that regard, detection of cell-secreted molecules is of paramount importance for observing the changes in tissue/organ construct upon certain stimulus. Thanks to advances in both microfluidics and sensing technologies, it is possible to build sensors integrated microfluidic platforms for detection of cell-secreted molecules. In this section, we will review current technologies available for the purpose of cell secretome monitoring.
Cytokines
Cytokines, signalling, low molecular weight soluble proteins secreted from both immune and non-immune cells possess important roles in immune responses such as proliferation, migration or activation of cells (107). In addition to that, cytokines were shown to play critical roles in cancer progression, cell-cell signalling and also tissue damage repair. Hence, development of sensors for cytokine detection is an obvious prerequisite for the study drugs influencing immune system, cell-cell signalling and cancer (108). The most widely used technique for cytokine detection is based on immunoassay such as in conventional ELISA assays, antibody array assays (109-111), bead-based assays (112) and aptamer-based assays (109,113,114).
In terms of cell culture and OOCs monitoring, cytokine sensors have been developed for monitoring liver cell signalling during injury (115), unravel the functional properties of individual immune cells (112), detection of activated T-cells (114), observe the secretion levels upon mitogenic stimulation (113).
As being highly thermally and chemically stable, aptamers have been preferred to antibodies for detection of protein-structured molecules. Several groups have used aptamers also for cytokine detection (109,113,114). Inflammatory cytokines, interferon gamma (IFN-γ) and tumor necrosis factor alpha (TNF-α) have been quantified via anthraquinone and methylene blue redox reporters labelled aptamers on Au electrodes (113). Different cytokine secretion levels of two cell types; primary human CD4 T-cells and U937 monocytic cells have been monitored concurrently over 2 h via microfluidic device. Similarly, another group have developed microfluidic co-cultures integrated with biosensors (Figure 6A) for continuous monitoring of TGF-β signalling in hepatocytes and stellate cell lines upon alcohol injury. The results showed that, designed microsystem enable monitoring paracrine crosstalk between two cell types communicating via transforming growth factor-beta (TGF-β) (115). Another advantage of the use of aptamers is that the sensor can be regenerated over saturation. Revzin et al. used this phenomenon to develop a reconfigurable platform with sensors and cells to monitor mitonegically activated T-cells via detection of interferon-γ (IFN-γ) by aptamer based regenerative electrochemical sensor. Sensors showed minimum loss over regeneration cycles and cells seem to be protected from regeneration solution, i.e., urea, thanks to microcups (Figure 6B) (114). The same group developed another cell-culture/biosensor platform with the same device, using the similar sensing approach for detection of TGF-β1 from activated stellate cells (Figure 6C) (109).
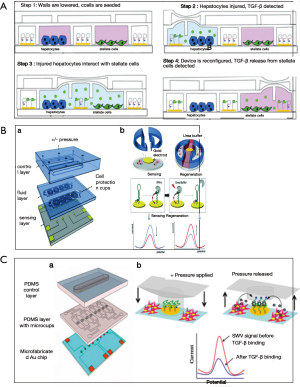
When it comes to single cell monitoring, and cytokine sensing from single cells, droplet based microfluidic approaches are taken into consideration since the method enables the manipulation of fluidic packets in the form of droplets and hence makes it possible to do high throughput monitoring (112). Huck et al. developed a droplet based microfluidic platform (Figure 7A) for secretion of two cytokines; IL-2, IFN-γ, TNF-α from single activated T-cells in droplets (112). Antibody-modified cytokine capture beads and single cells are encapsulated inside monodisperse agarose droplets to be able to detect the cytokines via measurement of the intensity of staining of the beads by flow cytometry (112). In a more recent work where again antibody coated microbeads were used to capture cytokines and monitor secretion levels of hepatocyte growth factor (HGF) and TGF-β1 over the course of 7 days. Developed microfluidic system enables local cytokine monitoring of primary hepatocytes that are separated from sensing chambers via permeable hydrogel barrier (Figure 7B) (116).
Other cell secretoms
Apart from cytokines, there are also various types of proteins, microRNAs and exosomes secreted by cells and gives hints about the fundamentals about cell biology for wide variety of applications including theraphy and diagnosis. For instance, albumin and glutathione-S-transferase-alpha (GST-α) and transferrin have been reported to be important biomarkers for liver. Hence researchers developed microfluidic platforms to detect secreted liver biomarkers via electrochemical sensors (9,72,117). A microfluidic magnetic bead-based EC immunosensor was developed to detect transferrin secreted by HepG2 cells cultured in bioreactors. Since the magnetic microbeads can be replaced with new ones after completing one cycle of measurement, the microelectrode reported to be usable over many measurements. The platform was used to monitor acetaminophen-induced toxicity over HepG2 cells (72). Another example on liver-on-a-chip platform monitoring aimed detection of albumin and GST-α via a label-free, regenerative electrochemical biosensor that enabled monitoring for a week of culture time (117). The same group further integrated those electrochemical sensors for monitoring dual-OOC system that consists of liver and heart in an automated manner (9).
Creatine kinase-MB (CK-MB) and troponins have been considered as cardiac damage or injury biomarkers. Therefore, researchers have developed sensing platforms for monitoring changes of those biomarkers for acute cardiac toxicity. A microfluidic aptamer sensor for detection of CK-MB has been designed to be able to monitor the changes in human embryonic stem cell-derived cardiomyocytes upon drug exposure (73).
Sensors for physical parameters
Sensors for physical parameters detection/control of system parameters
Main advantage OOCs stays in their ability to be highly micro-engineered. Indeed, different micro-sensors can be integrated for monitoring and controlling the micro-environmental conditions of the cultured cells (3). Thanks to continuous monitoring of physical parameters the maintenance of the physiological cell functions of cultured tissues and organs can be achieved. Moreover, since these physiological systems are characterized by metabolic reactions, it is imperative to control some crucial factors, among which pH, oxygen, temperature, glucose consumption and lactate production, to provide indication on the dynamic state of the human-on-a-chip system (118). Therefore, such systems should integrate sensing platforms for metabolic critical compounds, chemical signalling between organ compartments, fluid manipulation and microscopic investigation (119).
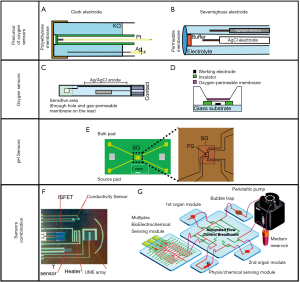
In particular, mitochondrial function plays a principal role in facilitating the cells growth and metabolism. When cells are exposed to an atmosphere with reduced oxygen concentration (hypoxia), cells adapt to loss of mitochondrial respiration by using anaerobic pathways such as glycolysis and glutaminolysis (120). This phenomena leads to undesirable stress in the cell culture, hence real-time measurement of dissolved oxygen (DO), even combined with sensors for pH, glucose and lactate, may preliminarily reveal and prevent from mitochondrial dysfunctions (121). Two types of oxygen sensors can be adopted for organ-on-a-chip applications: optical and electrochemical. The optical-transparency intrinsic of the microfluidic material enables a fast integration with optical sensors so that remote and non-invasive measurements are possible. On the other hand, the simplicity of electrochemical sensor architecture makes this method robust and reproducible for on-line monitoring (122). The first widespread DO sensor commonly adopted was proposed by Wolf et al., shown in Figure 8A (123). It consists of platinum working electrode (WE) as cathode and silver chloride reference electrode (RE) as anode able to detect the current from the oxygen reduction reaction (ORR):
O2 + 2H2O + 4e- ↔ 4OH-
O2 + 4H+ + 4e- ↔ 2H2O
A potentiostat is adopted to drive the measurements. It applies a stable potential of −700/−800 mV between WE and RE to make the reaction happen and to gather the current coming from it (128). The generation of electrons is proportional to the oxygen concentration (122). Major drawback comes from the formation of a diffusion layer nearby the surface of the electrode where oxygen is depleted. In this area, the cell may suffer from hypoxia leading to mitochondrial disorders. To overcome this limitation, to enhance the selectivity of the electrode and to avoid bio-fouling, a polytetrafluoroethylene (OTFE) membrane around WE can be adopted (129).
In order to achieve a constant pH it is needed to maintain equilibrium between atmospheric and dissolved CO2. Severinghaus, based on the R. W. Sow principle, realized the first CO2 sensor based on pH measurement, Figure 8B (123). The sensor consisted of a glass pH electrode arranged with a reference electrode in an electrolyte envelope on which the CO2 permeable membrane is mounted. Once the dissolved CO2 diffuses into the middle compartment of the electrode, a carbonic acid dissociation reaction happens (CO2 + H2O ⇌ H2CO3 ⇌ H+ + HCO3-) and the pH of the solution changes. The change in pH is sensed by the electrode and it is completely dependent on the pCO2. However, this indirect method to evaluate pCO2 is still unreliable since pH is affected also from other acidic species and not simply by CO2. Moreover, even if Severinghaus-kind electrodes were adopted for in-situ monitoring (130), they are bulky and difficult to be integrated within microfluidic systems. To this aim, there were many attempts for miniaturizing this kind of electrode. Suzuki et al. developed a miniaturized oxygen electrode, as shown in Figure 8C, and discussed the influence of cathode size on the data accuracy by testing seven different dimensions raged from 25×25 to 500×500 µm (124). The micro-square cathode and anode are realized as thin-film patterns on a glass substrate and are both immersed in the solution. A silicon rubber gas-permeable membrane was used. It was found that the main parameters to be affected by the cathode dimensions were: (I) flow dependence of the output current that increases with smaller dimensions; and response time that becomes shorter as the cathode becomes larger. Similarly, Wu et al. were able to miniaturize a Clark oxygen sensor and to integrate it with microstructures, can be seen in Figure 8D (125). It consists of a glass substrate with a three-electrode configuration and a PDMS oxygen-permeable membrane. By this sensor a fast response time of 6.8 s, a good linearity with a correlation coefficient of 0.995, and a long lifetime of at least 60 h.
Optical techniques are more advantageous in case of low oxygen levels since it does not cause the formation of the oxygen-depletion region around the electrode surface. Moreover, optical sensors do not need neither physical and electrical contact between the electrode inside the solution nor the detector (122). These sensors are based on photoluminescence quenching effect of oxygen that decreases the intensity/lifetime of luminophores. The degree of quenching depends from oxygen partial pressure. There are two main fluorophores to be chosen for their high quenching constants: ruthenium-based (131) and metalloporphyrin-based (132). Metalloporphyn-based dyes are preferred to avoid adding toxic material, as ruthenium is, into culture media. If the oxygen detection is based on the intensity of the luminescence effect, the set-up is simpler based on an emitted light with a wavelength to excite the luminophore. Unfortunately, this approach is susceptible to photobleaching, that decreases the intensity of luminescence after constant long-term excitation, to small movements in cells (133), to changes in optical geometry, optical properties and position of the sensor and to fluctuations in the light sources (122). These limitations are overcome by fluorescence lifetime imaging microscopy (FILM) that adopts modulated excitation source make detection in either time or frequency domain (134,135).
Finally, oxygen-sensing dyes can also be incorporated into fiber-optic probe. This configuration enables the possibility to move the probe within the sample while measuring (136). Another indicator for the respiratory system and alveoli functionality comes from CO2. Therefore, it is important to be considered especially in lung-on-a-chip studies. CO2 is produced during glucose consumption, and released into the blood, carried by blood to the lungs where it is exhaled. Since it is a weak acid, it changes the pH.
Therefore, other solutions were considered as ion-sensitive field-effect transistor (ISFET) and optical sensors. Optical sensors consist of optodes prepared by optically clear thin films attached covalently with fluorescent entities sensitive to pH (137). Hirura et al. fabricated a pH optode device by attaching a fluorophore agent with a PMS thin film (138). However, optodes suffer from small-range measurements (127) and ISFET are preferred technology to sense pH.
The original structure of the ISFET is characterized by a gate oxide that acts as sensing dielectric unit since it is in direct contact with the electrolyte solution (139). The voltage difference at the solid/liquid interface (∆ϕ) is converted in pH value by Nernst equation ∆ϕ= RT/F ln ai1/ai2; where R is the gas constant, T the absolute temperature and F the Faraday constant, ai = fi × ci are the ion activities and fi is the activity coefficient (in diluted electrolytes fi =1) (140). Moreover, considering the complexity of cellular reactions, it may not be sufficient to monitor just a single parameter. Therefore, a Physiocontrol-Microsystem (PCM) with various sensor elements, combined with small sized cell culture areas, able to measure respiration and acidification at the same time were introduced, for detailed schematic please see Figure 3 in (141). A single membrane-free device for on-line detection of both values at the same locus was manufactured with CMOS chip-technology (142). This method can be easily miniaturized to subcellular dimensions so that several sensors could be placed under one cell. The device designed by Krommenhoek et al. provides another example of integrated electrochemical sensor for measuring different parameters simultaneously, including pH and oxygen (127). While the oxygen concentration is measured by the UMEA, pH variations are registered through an ISFET. Both the sensors are included on the same device with also a thermistor to sense the temperature.
Another possibility is to modify the basic structure of the ISFET with extended gate (EG-ISFET), as in Figure 8E, where the sensing oxide is decoupled from the gate oxide by using an extended conductive layer (126). This more robust structure configuration where gate oxide is protected from the electrolyte solution ensures long time measurements inside the liquid.
Even smaller oxygen electrodes, represented in Figure 8F, were obtained by Krommenhoek et al. that realized an ultra-microelectrode array (UMEA) compatible with the 96-well plate format (127). Dissolved oxygen is measured amperometrically with an accuracy of ~0.2 mg/L.
Also it would be important to monitor osmolarity together with pH. Indeed, to ensure the normal development and life of cells, it is expected to keep both these variables unperturbed (143). The osmolality influences the cell viability and growth since it regulates the transport of water and nutrients through cell membranes while a constant pH ensures isotonicity of the culture (144). The most promising technique to monitor simultaneously pH and osmolality in cell culture is based on near-infrared spectroscopy (NIRS) technique, presented by Mattes et al. They addressed the in situ, real-time monitoring for both pH and osmolality parameters in bioreactors. In this way, a closed-loop culture feedback able to replace osmolality and pH measurements off-line or current on-line pH methods can be provided to ensure optimum cell performances and productivity.
Another way to sense osmolality relies on the non-selective transient receptor potential channel subfamily V member 4 (TRPV4). It was studied in cells from renal epithelium, its activation due to hyperosmotic stimuli, and it was characterized as a potential mechano-sensitive channel for flow and osmolality sensing (141).
The transparency properties of the micro-fluidic systems, not only allow the possibility to adopt optical techniques to sense biophysical parameters of the microenvironment (e.g., DO and pH), but also the detection of biochemical parameters such as cell/tissue viability (e.g., cell motility and live/dead assay) (145). Zhang et al. developed a miniature microscope from off-the-shelf components that can be mounted at the bottom of any microfluidic bioreactors and devices. The resolution is <2 µm and the adjustable magnifications of 8–60×.
In situ microscope is another technique adopted in cell culture to acquire images inside bioreactors (in-situ) during long-term perfusion. This technique allows images of cells in a defined volume inside a bioreactor without sampling or bypass construction. Therefore, the system can be cleaned and serviced without interruption of the process or risking contamination (146). Not only 2D in situ imaging has been performed but also 3D, under static and dynamic flow conditions. This aids in identification and optimization of the microenvironment properties and flow conditions that enhance cell response (147).
Another possibility is to build the optics out of the same fluidic toolkit, in this case an optofluidic device is realized (148). This solution is attractive but still under investigation. Furthermore, microscopy can be paired with spectrometry analysis. Impedance spectroscopy is adopted to study the dielectric characteristics of particles and cells in a label-free manner. It enables cell counting, size quantification, and cell classification and phenotype characterization in continuous flow (149). Recently, Wang et al. realized for the first time a cell/particle position detection technology using a single-channel impedance spectroscopy device with low-cost and high-throughput (150).
As concerning organ-on-a-chip systems application, only few examples of full-integration platforms are reported in literature. Zhang et al. we reported a fully integrated modular physical, biochemical, and optical sensing platform, interfaced through a fluidics-routing breadboard with a multi-organ-on-a-chip system to achieve in situ, on-line, and automated sensing of microenvironment biophysical and biochemical parameters (9). The system is visible in Figure 8G.
Sensors for monitoring cell fate
Cells in their native environment are exposed to multiple cues that vary in time and space, including gradients of cytokines and secreted proteins from neighboring cells, biochemical and mechanical interactions with the ECM, and direct cell-cell contacts. These cues cannot easily be achieved by standard tissue culture. However, microfabricated systems can mimic cells with these cues in a manageable and reproducible mode that can be used to link cell culture with integrated analytical devices. This integration can be verified for investigation of the biochemical processes that govern cell behavior. In this part of the review paper, we will discuss more about sensors for investigating the fate of the cells, cell cycle analysis, cell viability, single cell analysis, differentiation analysis, cell sorting, and cell separation.
A complex chemical and mechanical microenvironment, which is composed of different secreted factors, extracellular matrix, and direct interactions with other cells, is needed to direct and control the stem cell renewal and lineage fate. Therefore, it is crucially important to have reliable and sensitive platforms to be able to constantly monitor the artificial tissue environment in terms of various physiologically relevant parameters to evaluate the functionality of tissue-engineered structures. These parameters and signals can be mimicked and precisely controlled in a high throughput automated manner and integrated several functions using microfluidic portable platforms at microscale (151,152). So, one of the key challenges for researches in tissue engineering is the development of biosensors for point-of-care (POC) applications and combining it with microfluidic platforms, as many biomarkers have to be monitored to evaluate the functionality of any tissue engineered construct in vitro (22). Monitoring the transmission of various physical and chemical signals, such as changes in oxygen consumption, pH, membrane potentials, ion concentrations, and release of various metabolic compounds and proteins in living cells can give insights into cellular activities in real time (22,153).
Each stage of cell adhesion i.e., attachment, spreading, migration, and immobilization, activates signals that regulate cell survival, growth, cell-cycle progression and differentiation, and motility in multiple cell systems and involves changes in cell morphology and cytoskeletal structure (154). Biosensors have a crucial impact on tissue engineering applications where concentrations of different biomolecules such as glucose, adenosines, and hydrogen peroxide levels play important roles in determining the fate of the cells and tissues, especially in maintaining three dimensional cell cultures and developing “OOCs” models (22).
Conventional molecular biosensors usually use specific chemical properties or molecular recognition mechanisms to identify a particular, singular agent, and thus are limited to detecting only known bio-threats. There are several advantages of cell-based sensors in comparison to conventional molecular sensors such as the ability to detect many forms of substances, including chemical toxins, bacteria, and viruses, a high sensing efficiency and rapid response, they can provide insights into the physiological effect of an analyte and they are low cost, all because of the fact that the sensing receptors are embedded in cell membranes and the protein separation and purification is not necessary (154).
Cell-based sensors have different applications including pH detection to monitor metabolism activity, fluorescence imaging of protein-expression related to cell signalling, electrical probing and analysis of intra and extra-cellular membrane potential using microelectrodes, and impedance detection for monitoring cell adhesion and spreading.
One of the most important cell-based biosensors is impedance biosensors, which are typically developed by immobilizing a group of cells on an array of electrodes on an insulative substrate and is integrated with microelectronics for real-time data acquisition, analysis, and display. Dielectric spectroscopy as viable biosensing tool was reviewed elsewhere (155) where also common ECIS measurement techniques using microelectrode arrays with the respective sensing and counter electrodes, and interdigitated electrode design were depicted in a figure [see Figure 5 in (155)]. Figure 9A depicts a typical flow cytometer with optical detection system (156). Design of a microelectrode array for cell impedance sensing was shown by Figure 9B (155).
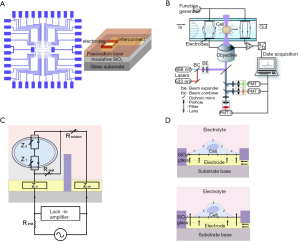
The impedance model of the cell adherence to the electrode and its schematic representation was illustrated by Figure 9D and Figure 9E respectively (154). By monitoring electric changes at the contact between the cell and electrode, cellular impedance biosensors are capable of determining the events of cell adhesion, spreading, growth, motility, and death for any adherent cell types (both excitable and non-electrically active) (154). The cytotoxicity of various agents such as chemicals, drugs, cell tags, and implants, along with allowing the observation of the effect of various stresses on cells can be screened using impedance measurements. Therefore, it can replace slower and invasive traditional cytotoxicity assays (155).
Sensors for cell viability
The viability of cells attached to the substrate is essential for the cells to function as sensing elements. The cell’s dye can be either pathological or programmed death. Programmed cell death or Apoptosis, which is the desired response of a cancerous tumor to effective chemotherapies or radiation treatments, is characterized with dramatic changes in cell morphology, ionic channel conductance, and extracellular membrane integrity, as well as altered intracellular structure. These electrical property changes can be diagnosed quickly and inexpensively by cell impedance biosensors at significantly reduced cost and expedited assessment procedure, and thus these biosensors offer an optimal approach in anti-cancer drug screening (154). This technology can improve healthcare in the developing world and provide rapid diagnoses for livestock on isolated farms, as integrated chips using impedance can eventually reach clinical settings and will be useful in remote regions that lack good infrastructure or experienced personnel (155).
A study focused on monitoring the apoptosis-induced changes in cell shape in an integral and quantitative fashion with a time resolution in the order of minutes using ECIS. It has been shown that the response to an apoptosis which was induced by cycloheximide (CHX) and verified by biochemical and cytological assays resulted in a rapid and monotonic decrease in impedance (157).
Sensors for single cell analysis
One way to perform impedance for single-cell analysis is to use smaller microelectrode arrays to reduce the size of sense electrodes so that only a single cell fits on them. Typical microelectrode array setups contain several cell-sized sensing electrodes which are coated with a self-assembled monolayer and a peptide to attract single cells to the electrode and one or more large counter electrodes (158,159). Another single-cell system uses a flow bottleneck to capture cells individually, and the electrodes are situated at the bottleneck in order to take impedance measurements while the cells are trapped which can then be released upon the application of a high fluid pressure (160). One of the other single cell platforms use scanning dielectric microscope in which an electrode probe is mechanically scanned over a cell (161). Another platform that can be conducted for cell clusters, and single cells uses a light addressable impedance detection chip in which a laser selects and enables measurement of cells (155).
Moreover, impedance flow cytometry (IFC) is capable of measuring the electrical properties of single cells. IFC technology uses direct current (DC) or low frequency alternate current (AC) for impedance measurements. Impedance measurements provide information about membrane capacitance, cytoplasm conductivity, and cytoplasm permittivity as a function of frequency in the range from 40 Hz to 1 GHz.
To improve the efficiency of these devices, they have to be sensitive, stable and reproducible to detect very low quantity of any toxic substance in the cellular environment. The ECIS system is the most common impedance cytosensor, which can be used for characterization of cell attachment, micromotion, and cytotoxicity (154). In ECIS, adherent cells were cultured on an array of eight small gold electrodes and the impedance is changed based on the behavior of the cells. For instance, it increases because the adhesion of the cells to the substrate, or the measured impedance continued to fluctuate to reflect the constant motion and metabolic activity of the cells after the cells were fully spread. Also the impedance is changed by stimulation of the cells with external chemicals. To demonstrate the cell motility and mortality, the quantitative data are taken in real time and in a continuous fashion (162).
Blood glucose monitoring has been established as a valuable tool in tissue engineering applications, as it is a critical indicator of metabolic activities of cells (163,164). Since maintaining normal blood glucose levels is recommended, a number of different biosensing approaches including electrochemical biosensors that are frequently used for glucose oxidase or glucose dehydrogenase detection, optical biosensors for glucose detection using inactive apoenzymes, binding proteins, and receptors and also nanobiosensor have been developed for glucose monitoring in engineered tissue constructs in real time during their fabrication, proliferation, and growth, which can give the consumption of glucose by the cells (22,165-167).
Sensors for cell differentiation analysis
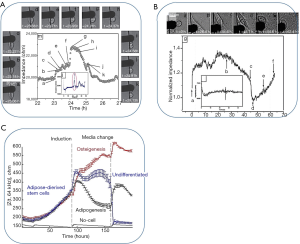
One of the most important applications of sensors in developmental biology is the evaluation of different stages of cell locomotion, cell-surface interaction since they are extremely important for cell differentiation. The differentiation of the cells is studied in a wide range of applications like studying the cancer cells, screening the drugs, disease modelling, and developmental biology researches. To better understand the impact of different factors on cell fate, cell differentiation sensors can be useful and informative. Cell differentiation can be characterized by changes in cell motility, cell morphology, aggregation, cell adhesion, ion channel activity, gene and protein expression, etc. which leads to a dramatic change in the overall impedance spectrum of the cells. Different stages of cell mitotic divisions as well as cell motility were successfully probed by positive dielectrophoretic device for mammalian cells (168). Figure 10A shows the impedimetric changes of a single cell during its mitotic divisions. As can be seen from that figure, the impedance values decreased between periods shown by G and K which corresponds to anaphase, telophase and cytokinesis whereas the period till the peak point, A to G, shows the cells morphological changes from flattened to spherical during prophase as well as condensation of chromosomes during metaphase (168). The same sensor was also used for measuring cell motility (168) by recording changes in impedance values for two cells initially attracted at the edge of the electrode. By looking at Figure 10B, one can understand that, attachment (snapshots A-B), detachment (snapshots B-D) and re-attachment (snapshots B-D) of cells brings about significant changes on impedance values (168). For instance, in a study, the adipose-derived stem cells (ADSCs) were induced into the differentiation towards adipocytes and osteoblasts (169). Four days after induction towards differentiation, the cell membrane capacitance (Cm) values have changed intensely as can be seen from Figure 10C from Cm =2.25±0.27 µF/cm2 to Cm =1.72±0.10 µF/cm2 to for adipo-induced cells to osteo-induced cells (169). Impedance measurements have also been conducted for investigation of the impact of environmental factors such as cell niche (170), or toxic components like pesticide (171) on the differentiation of the cells. Figure 10C shows the variation of impedance based on the differentiation of the cells. Real-time and label-free monitoring of ADSCs induced toward osteoblasts and adipocytes. (A) Time-course measurement of mean impedance, |Z(t,f)|, at 64 kHz, for different groups throughout early.
In tissue engineering, the microporous polymer-based scaffolds are widely used for culturing and proliferating cell within entire volumes of a predefined shape. However, it is difficult to assess directly the in vitro cellular proliferation and differentiation occurring within 3D scaffold constructs (155). Therefore, a viable monitoring technique within macro chambers, using FEA modelling techniques and the implementation of cell model and thin layer approximations, coupled with parallel-plate platinum electrodes, has been successfully conducted for the non-invasive monitoring of 3D cellular clusters and associated epithelial cell differentiation processes.
Other factors which have an important role in a wide variety of different biological responses such as apoptosis, cell proliferation, migration and differentiation, cytokine release, and necrosis are the extracellular ATP, ADP, and uridine triphosphate (UTP). Llaudet and coworkers developed a microelectrode to be placed inside the tissue for in vivo measurements of ATP. In another research, a biosensor has been developed, which can be placed near the ATP-releasing target cells. This technique gives an accurate result of the extracellular ATP concentration. Recently, Xie and coworkers developed a novel localized surface plasmon resonance (LSPR) array chip for facile, label-free, and high throughput detection of ATP using a normal microplate reader which can be used for miniaturized and high throughput detection of biological samples in tissue engineering applications (172).
A multiparametric micopysiometry platform has been developed to monitor the metabolism of T98G human brain cancer cells cultured in dynamic flow conditions (102). Several microfabricated biosensors were integrated in the cell chamber and the levels of pH, oxygen consumption, and the production of cell metabolites were monitored by using external equipment (e.g., potentiostat). Similarly, Hu et al. included a light-addressable potentiometric sensor (LAPS) in a microfluidic system to monitor the metabolism of human breast cancer cells in real time. Microheaters and micropumps were also integrated to these systems for controlling the temperature and handling different fluids (9,173).
Another type of biosensors are potentiometric biosensors which allow non-invasive, real time monitoring of the extracellular environment changes by measuring the potential at cell/sensor interface to observe the overall cell cytotoxicity. Potentiometric biosensors can measure many processes in living cells, which have electrochemical characteristics. Wang et al. employed a potentiometric sensor array to investigate the cytotoxicity of hydroquinone to cultured mammalian V79 cells. The results showed that hydroquinone exposure affected cell proliferation and delayed cell growth and attachment in a dose-dependent manner. The potentiometric biosensor provides non-invasive and real time monitoring of the cellular reactions and also is more sensitive for in vitro cytotoxicity study which makes it more promising system for drug discovery compared with traditional methods (153).
Sensors for cell sorting, cell separation
Monitoring the cell cycle behavior of individual cells separately, has an imperative role in developmental biology due the fact that it can facilitate the understanding of developmental processes such as pattern formation, morphogenesis, cell differentiation, growth, cell migration, and cell death. It has also crucial impact on cancer researches, especially cancer-on-chip studies to investigate the fate or metastasis of the cells. The next generation of OOC devices will be designed for disease modeling such as cancer and genetic disorders, which require the very precise cell studies in cellular level. Therefore, combining the sensors for cell sorting and cell separation in the designed chips are imperative to investigate the alterations in cell morphology and gene expression due to metabolic pathways or effect of drugs and to purify the population of the cells to have a better understanding of the changes that will occur.
The development of numerous microscale separation techniques is a result of need for efficient, accurate and high throughput cell separation, an essential preparatory step in many biological and medical assays. Microfluidics-based sorting in comparison with conventional methods provides numerous advantages, including reducing sample volumes, faster sample processing, high sensitivity and spatial resolution, low device cost, reducing the size of necessary equipment, eliminating potentially biohazardous aerosols, simplifying the complex protocols commonly associated with cell sorting and increased portability (174). The techniques for cell separation can be divided into two main groups: active systems which generally use external fields such as acoustic, electric, magnetic, and optical; and passive systems which use inertial forces, filters, and adhesion mechanisms to purify cell populations (175). Separation of the cells has an important role in the early diagnosis of cancer. There are a number of molecular markers for cancer diagnosis and these can include proteins, peptides, over/under expression of gene markers and gene mutations (176).
Label-free cell sorting in microfluidic devices relies on the physical differences in the properties of cells such as size, shape, density, elasticity, polarizability, and magnetic susceptibility instead of solely relying on surface markers for labelling cells with fluorophores or beads. Label-free sorting generally requires the least amount of preparation, making it a highly attractive option for cell sorting (175). Blood analysis is one of the applications of lab-on-a-chip types of devices which takes advantage of one of the physical properties of the blood, which is the blood cells’ heterogeneity in parting (177). Some different approaches for sorting the cells have been shown in Figure 11, first row. Figure 11A shows a spiral microfluidic channel to apply centrifugal forces to focus and sort cells by size (178). The equilibrium position of any particle at various sizes depends on the ratio of lift (FL) and dean drag forces (FD) that varies to the particle diameter in third power the particles equilibrating them into focused streams around the microchannel perimeter (178).
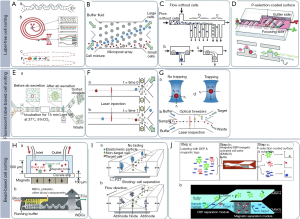
Figure 11B, provides deterministic lateral displacement which is an important example of navigating the cells through an array of posts for sorting by size. Design of the array features can control over sorting such that cells smaller than a critical radius (a < Rc) move with the convective flow and cells larger than a critical radius (a > Rc) move in a direction dictated by the arrays (179). Hydrodynamic filtration (Figure 11C) is another type of microfluidic filtration in which aligned cells are separated by multiple branched outlets, whereby the fluid draining from the outlets pulls cells from the walls of the main channel at rates that scale according to their size (180). Another type of cell sorting can be done by deterministic cell rolling (Figure 11D). In this case, target cells interact with the surface, roll across the ridges, and laterally displace toward the gutter side whereas non-target cells flow over the ridges, not interacting with the surface, and exit on the focusing side (181).
Currently, one of the most useful approaches for sorting the cells is fluorescent label-based cell sorting, as its intensity is very sensitive. The parameters that can be measured by fluorescence are temperature, cell function, flow velocity, flow profiles and polymer dynamics (177). Many research groups have used fluorescent labels to identify cells in the microfluidic regime for sorting by a variety of mechanisms, such as electrokinetic mechanisms, acoustophoresis, optical manipulation and mechanical systems. This approach is similar to FACS devices, which generally activate by ordering cells in flow streams for sorting. Since each cell is processed discretely, fluorescent label-based approaches are often associated with high efficiencies, which makes it the mainstays of modern cell sorting technologies and a viable option for many microchip cell-sorting devices. However, this method has some drawbacks such as the inconvenience of serial detections, discrete sorting, and the inability of the fluorescent labels to directly contribute to the sorting process which has led to the development of other effective techniques for rapid cell sorting, including bead-based and label-free cell sorting systems (175). The middle row of Figure 11 shows some examples of fluorescent label-based cell sorting. Figure 11A shows that cells compartmentalized into emulsions with beads coated with capture antibodies can be used to analyze the secretion of antibodies from cells for downstream sorting using DEP (182).
Electroosmotic flow (Figure 11B) is based on the movement of a fluid due to the electrically induced migration of solvated ions which leads to transporting cells suspended within the fluid (183). Figure 11C indicates the cell sorting by optical force switching. Cells will deflect when scattering forces (FS) exceed gradient forces (FG) from a focused laser beam; however, when FG exceeds FS, cells are optically trapped. In this figure, a hydrodynamically focused stream of cells is shown which is aligned toward the waste outlet whereupon cells of interest detected by laser inspection are captured and displaced by optical tweezers for sorting (184). Another example to fluorescent label-based cell sorting is proposed by Lopez et al. (185) who used a microfabricated cantilever beam that is shifted from a ‘down’ position to an ‘up’ position when a corresponding pair of electrodes is activated to generate bubbles via isothermal electrolysis, which in turn exerts a mechanical force on the T-switch to redirect fluid flow [See Figure 5 in (185)].
Using particles of a specific material, size, and surface-binding capacity, bead-based cell sorting systems capture target cells, or sometimes non-target cells, for sorting in an external field. Based on the attachment of beads, bound complexes experience a force that is different from unbound cells. This method has the ability to sort cells at potentially faster rates and without large volumes of diluent. This method of cell isolation was soon miniaturized to the microfluidic domain to sort cells with magnetic and other types of particulate labels. Cell sorting by magnetophoresis (MAP) is simplified by the use of permanent magnets or electromagnetic coils to exert forces on cells labelled with magnetic particles, magnetically responsive cells, or cells suspended in ferrofluid. This approach is usually conducted to either de-bulk cell populations or to rare cells separation from native biological fluids.
Three ways of bead-based cell sorting has been shown in the last row of Figure 11. Magnetic isolation of CTCs in microfluidic devices (Figure 11A) are shown to capture and isolate CTCs under free flow in a magnetic field. In this approach, deterministic lateral displacement for filtering magnetically labeled CTCs and leukocytes and inertial focusing for focusing and sorting in a magnetic field are also used (185,186). Figure 11B shows acoustofluidic systems. This method of cell confinement can enable rapid, continuous, and discriminate sorting in which unlabeled cells focus at the acoustic node(s) and labeled cells focus at the acoustic antinodes for downstream collection. Using silicone elastomeric particles synthesized from nucleation and growth techniques, biofunctionalized elastomeric particles decorated with streptavidin can capture cells displaying biotinylated antibodies and confine those cells to the antinodes of an acoustic standing wave (187). Moreover, Figure 11C is a schematic of multi-target cell sorting using DEP and magnetic trapping. In this case, target cell A is captured with polystyrene beads depicted in green and target cell B is captured with superparamagnetic beads depicted in red. Target cell A is removed from non-target cells by dielectrophoretic forces and target cell B is removed from non-target cells by magnetic trapping (188).
Future outlook
Undoubtedly, OOC platforms are considered to be very promising tools for replacement of animal testing to perform drug toxicity studies as well as performing personalized medicine research. As well as increasing cost of drug development, rising ethical concerns on animal experiments result in the urgent demand of a valid human-on-a-chip model. However, current models of OOC provide relatively simple cell culture protocols or variety of organ models that often do not sufficiently present real human physiology or even organ functions. Among several challenges to design the valid OOC model, one of the most important one is to grow different cell types together in a 3D organ format. Another big challenge having build such an OOC model is to assess the functionality of this platform via monitoring of pathophysiological cell-cell interactions, cell secreted molecules and physical parameters of the culture microenvironment. To achieve this goal, sensors as well as microscopes that enable real-time and long-term monitoring should be integrated to OOC platforms. As pharmaceutical companies need high-throughput screening to be able to do profiling and pharmacokinetic evaluation, OOC are expected to have integrated sensors that enables medium to high-throughput monitoring in a sensitive and selective manner. Although most of the OOC lack in vasculature and high-throughput monitoring, thanks to advanced manufacturing technologies and microscale size of microfluidic devices that reduce the reagents volume and speed up the assays that require thermocycling, in future, OOC could be expected to meet with the needs of pharma to screen thousands of drug candidates.
As it is detailed in this paper, there has been growing interest in recent years in sensor development for OOC monitoring. However, there is still room for improvement on more sophisticated sensor design and integration. One of the most urgent needs in terms of sensor design is real-time and continuous monitoring. Even though optical and electrochemical approaches are suggested as real-time monitoring alternatives, sensor saturation and regeneration is a big issue. Various regeneration techniques have been developed to tackle this issue but none of the designed sensor managed to monitor any OOC platform more than a week. Aptamers shown to hold great promise due to their chemical stability over regeneration (9,73,113,114) and providing highly-sensitive new devices (189), 16, 4,472–4,476. In addition to aptamers, bead-based assays could be alternative solution to long-term monitoring since the used ones can be discharged (72,116). Another challenge regarding sensors is automation, which could have been achieved in only a few papers. Automation of sensors and data transmission would enable translation of such platforms to Clinique since it would replace labor-intensive and timely operations. To satisfy the regularly authorities’ needs on drug testing, medium to high throughput sensors and monitoring strategies should be incorporated to OOCs. In that respects, single-cell barcode chips seem to be the strongest alternative by enabling quantification of numerous protein concentrations with immuno-sandwich assays via spatially encoded antibody barcode (110,111). In terms of cell fate monitoring, MEAs based on impedance sensing provides very valuable information and could be improved in future to do real-time 3D mapping and manipulation of electrical activity. In addition to microfabrication, materials science and microfluidics technologies molecular biology and proteomics studies are indispensable for sensor design since new class of biomarkers are always needed to assess the functionality of OOC platforms.
Development of physiologically relevant OOC systems to be used for medical or basic science investigation purposes requires extensive integration of a wide variety of disciplines from molecular biology to microfabrication. To achieve “clinical trials on chip” and to proceed with personalized medicine, OOCs possess a big responsibility and challenges to tackle with. Sensor integration in that end is one of the “musts” to be taken into consideration. Multidisciplinary collaboration, not only with different disciplines but also with pharmaceutical companies and regulatory authorities is needed to design sensors that are capable of assessment of OOC platforms in a real-time, continuous and high-throughput manner.
Acknowledgments
Funding: This project has received funding from the European Union’s Horizon 2020 research and innovation programme funder the Marie Skłodowska-Curie grant agreement No. 665667 and partial funding from SNF Multidisciplinary CR32I3/156915.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/mps.2018.01.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Khademhosseini A, Langer R. A decade of progress in tissue engineering. Nat Protoc 2016;11:1775-81. [Crossref] [PubMed]
- Huh D, Matthews BD, Mammoto A, et al. Reconstituting Organ-Level Lung Functions on a Chip. Science 2010;328:1662-8. [Crossref] [PubMed]
- Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat Biotechnol 2014;32:760-72. [Crossref] [PubMed]
- Kim HJ, Ingber DE. Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr Biol (Camb) 2013;5:1130. [Crossref] [PubMed]
- Giese C, Lubitz A, Demmler CD, et al. Immunological substance testing on human lymphatic micro-organoids in vitro. J Biotechnol 2010;148:38-45. [Crossref] [PubMed]
- Korin N, Kanapathipillai M, Matthews BD, et al. Shear-Activated Nanotherapeutics for Drug Targeting to Obstructed Blood Vessels. Science 2012;337:738-42. [Crossref] [PubMed]
- Wagner I, Materne EM, Brincker S, et al. A dynamic multi-organ-chip for long-term cultivation and substance testing proven by 3D human liver and skin tissue co-culture. Lab Chip 2013;13:3538. [Crossref] [PubMed]
- Maschmeyer I, Lorenz AK, Schimek K, et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip 2015;15:2688-99. [Crossref] [PubMed]
- Zhang YS, Aleman J, Shin SR, et al. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc Natl Acad Sci U S A 2017;114:E2293-302. [Crossref] [PubMed]
- Maoz BM, Herland A, Henry OYF, et al. Organs-on-Chips with combined multi-electrode array and transepithelial electrical resistance measurement capabilities. Lab Chip 2017;17:2294-302. [Crossref] [PubMed]
- Gaio N, Van Meer B, Quiros Solano WF, et al. Cytostretch, an Organ-on-Chip Platform. Micromachines 2016;7:1-14. [Crossref]
- Engle SJ, Puppala D. Integrating Human Pluripotent Stem Cells. Cell Stem Cell 2013;12:669-77. [Crossref] [PubMed]
- Wu MH, Huang SB, Lee GB. Microfluidic cell culture systems for drug research. Lab Chip 2010;10:939-56. [Crossref] [PubMed]
- van der Meer AD, van den Berg A. Organs-on-chips: breaking the in vitro impasse. Integr Biol (Camb) 2012;4:461-70. [Crossref] [PubMed]
- Adler S, Basketter D, Creton S, et al. Alternative (non-animal) methods for cosmetics testing: current status and future prospects-2010. Arch Toxicol 2011;85:367-485. [Crossref] [PubMed]
- Huh D, Hamilton GA, Ingber DE. From 3D cell culture to organs-on-chips. Trends in Cell Biology 2011;21:745-54. [Crossref] [PubMed]
- Verhulsel M, Vignes M, Descroix S, et al. Biomaterials A review of microfabrication and hydrogel engineering for micro-organs on chips. Biomaterials 2014;35:1816-32. [Crossref] [PubMed]
- Ghaemmaghami AM, Hancock MJ, Harrington H, et al. Biomimetic tissues on a chip for drug discovery. Drug Discovery Today 2012;17:173-81. [Crossref] [PubMed]
- Luni C, Serena E, Elvassore N. Human-on-chip for therapy development and fundamental science. Curr Opin Biotechnol 2014;25:45-50. [Crossref] [PubMed]
- Gradient B, For H. NIH Public Access. 2011;88:899-911.
- Agarwal A, Goss JA, Cho A, et al. Microfluidic heart on a chip for higher throughput pharmacological studies. Lab Chip 2013;13:3599-608. [Crossref] [PubMed]
- Hasan A, Nurunnabi M, Morshed M, et al. Recent advances in application of biosensors in tissue engineering. Biomed Res Int 2014;2014:307519 [Crossref] [PubMed]
- Bhise NS, Manoharan V, Massa S, et al. A liver-on-a-chip platform with bioprinted hepatic spheroids. Biofabrication 2016;8:014101 [Crossref] [PubMed]
- Lee H, Cho DW. One-step fabrication of an organ-on-a-chip with spatial heterogeneity using a 3D bioprinting technology. Lab Chip 2016;16:2618-25. [Crossref] [PubMed]
- Banaeiyan AA, Theobald J, Paukstyte J, et al. Design and fabrication of a scalable liver-lobule-on-a-chip microphysiological platform. Biofabrication 2017;9:015014 [Crossref] [PubMed]
- Khazali AS, Clark AM, Wells A. A Pathway to Personalizing Therapy for Metastases Using Liver-on-a-Chip Platforms. Stem Cell Rev 2017;13:364-80. [Crossref] [PubMed]
- Khetani SR, Bhatia SN. Microscale culture of human liver cells for drug development. Nat Biotechnol 2008;26:120-6. [Crossref] [PubMed]
- Prodanov L, Jindal R, Bale SS, et al. Long-term maintenance of a microfluidic 3D human liver sinusoid. Biotechnol Bioeng 2016;113:241-6. [Crossref] [PubMed]
- Zakhariants AA, Burmistrova OA, Shkurnikov MY, et al. Development of a Specific Substrate-Inhibitor Panel (Liver-on-a-Chip) for Evaluation of Cytochrome P450 Activity. Bull Exp Biol Med 2016;162:170-4. [Crossref] [PubMed]
- Ware BR, Khetani SR. Engineered Liver Platforms for Different Phases of Drug Development. Trends Biotechnol 2017;35:172-83. [Crossref] [PubMed]
- Meli L, Barbosa HSC, Hickey AM, et al. Three dimensional cellular microarray platform for human neural stem cell differentiation and toxicology. Stem Cell Res 2014;13:36-47. [Crossref] [PubMed]
- Choi SH, Kim YH, Quinti L, et al. 3D culture models of Alzheimer’s disease: a road map to a “cure-in-a-dish”. Mol Neurodegener 2016;11:75. [Crossref] [PubMed]
- Dollé JP, Morrison B 3rd, Schloss RS, et al. Brain-on-a-chip microsystem for investigating traumatic brain injury: Axon diameter and mitochondrial membrane changes play a significant role in axonal response to strain injuries. Technology 2014;2:106. (Singap World Sci). [Crossref] [PubMed]
- Pandey PK, Sharma AK, Gupta U. Blood brain barrier: An overview on strategies in drug delivery, realistic in vitro modeling and in vivo live tracking. Tissue Barriers 2015;4:e1129476 [Crossref] [PubMed]
- Doryab A, Amoabediny G, Salehi-najafabadi A. Advances in pulmonary therapy and drug development : Lung tissue engineering to lung-on-a-chip. Biotechnol Adv 2016;34:588-96. [Crossref] [PubMed]
- Grosberg A, Alford PW, McCain ML, et al. Ensembles of engineered cardiac tissues for physiological and pharmacological study: heart on a chip. Lab Chip 2011;11:4165-73. [Crossref] [PubMed]
- Aubin H, Nichol JW, Hutson CB, et al. Directed 3D cell alignment and elongation in microengineered hydrogels. Biomaterials 2010;31:6941-51. [Crossref] [PubMed]
- Giridharan GA, Nguyen MD, Estrada R, et al. Microfluidic cardiac cell culture model (µCCCM). Anal Chem 2010;82:7581-7. [Crossref] [PubMed]
- Selimović S, Dokmeci MR, Khademhosseini A. Organs-on-a-chip for drug discovery. Curr Opin Pharmacol 2013;13:829-33. [Crossref] [PubMed]
- Zhang YS, Arneri A, Bersini S, et al. Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials 2016;110:45-59. [Crossref] [PubMed]
- Marsano A, Conficconi C, Lemme M, et al. Beating heart on a chip: a novel microfluidic platform to generate functional 3D cardiac microtissues. Lab Chip 2016;16:599-610. [Crossref] [PubMed]
- Xiao S, Coppeta JR, Rogers HB, et al. A microfluidic culture model of the human reproductive tract and 28-day menstrual cycle. Nat Commun 2017;8:14584. [Crossref] [PubMed]
- Li WX, Liang GT, Yan W, et al. Artificial Uterus on a Microfluidic Chip. Chinese J Anal Chem 2013;41:467-72. [Crossref]
- Chang KW, Chang PY, Huang HY, et al. Womb-on-a-chip biomimetic system for improved embryo culture and development. Sensors Actuators B 2016;226:218-26. [Crossref]
- Ryu Hyunryul, Oh Soojung, Lee Hyun Jae, et al. Engineering a Blood Vessel Network Module for Body-on-a-Chip Applications. J Lab Autom 2015;20:296-301. [Crossref] [PubMed]
- Zheng W, Jiang B, Wang D, et al. A microfluidic flow-stretch chip for investigating blood vessel biomechanics. Lab Chip 2012;12:3441. [Crossref] [PubMed]
- Barkefors Irmeli. Sara Thorslund FN and JK. A fluidic device to study directional angiogenesis in complex tissue and organ culture models. R Soc Chem 2009;9:485-624.
- Zheng Y, Chen J, Craven M, et al. In vitro microvessels for the study of angiogenesis and thrombosis. Proc Natl Acad Sci 2012;109:9342-7. [Crossref] [PubMed]
- Buehler SM, Stubbe M, Bonk SM, et al. Cell Monitoring and Manipulation Systems (CMMSs) based on Glass Cell-Culture Chips (GC3s). Micromachines 2016;7:106. [Crossref]
- Nguyen TA, Yin TI, Reyes D, et al. Microfluidic Chip with Integrated Electrical Cell-Impedance Sensing for Monitoring Single Cancer Cell Migration in Three-Dimensional Matrixes. Anal Chem 2013;85:11068-76. [Crossref] [PubMed]
- Cho S, Islas-Robles A, Nicolini AM, et al. In situ, dual-mode monitoring of organ-on-a-chip with smartphone-based fluorescence microscope. Biosens Bioelectron 2016;86:697-705. [Crossref] [PubMed]
- Vala M, Robelek R, Bocková M, et al. Real-time label-free monitoring of the cellular response to osmotic stress using conventional and long-range surface plasmons. Biosens Bioelectron 2013;40:417-21. [Crossref] [PubMed]
- Prieto JL, Su H-W, Hou HW, et al. Monitoring sepsis using electrical cell profiling. Lab Chip 2016;16:4333-40. [Crossref] [PubMed]
- Shevchenko Y, Camci-Unal G, Cuttica DF, et al. Surface plasmon resonance fiber sensor for real-time and label-free monitoring of cellular behavior. Biosens Bioelectron 2014;56:359-67. [Crossref] [PubMed]
- Fang Y, Ferrie AM, Fontaine NH, et al. Resonant Waveguide Grating Biosensor for Living Cell Sensing. Biophys J 2006;91:1925-40. [Crossref] [PubMed]
- Zhang X, Wang T, Wang P, et al. High-Throughput Assessment of Drug Cardiac Safety Using a High-Speed Impedance Detection Technology-Based Heart-on-a-Chip. Micromachines 2016;7:122. [Crossref]
- Conant G, Lai BFL, Lu RXZ, et al. High-Content Assessment of Cardiac Function Using Heart-on-a-Chip Devices as Drug Screening Model. Stem Cell Rev 2017;13:335-46. [Crossref] [PubMed]
- Zhou J, Niklason LE. Microfluidic artificial “vessels” for dynamic mechanical stimulation of mesenchymal stem cells. Integr Biol (Camb) 2012;4:1487-97. [Crossref] [PubMed]
- Sei Y, Justus K, Leduc P, et al. Engineering living systems on chips: From cells to human on chips. Microfluid Nanofluidics 2014;16:907-20. [Crossref]
- Kuczenski B, Ruder WC, Messner WC, et al. Probing cellular dynamics with a chemical signal generator. PLoS One 2009;4:e4847 [Crossref] [PubMed]
- Chen Y-A, King AD, Shih HC, et al. Generation of oxygen gradients in microfluidic devices for cell culture using spatially confined chemical reactions. Lab Chip 2011;11:3626. [Crossref] [PubMed]
- Elzinga K, Tyreman N, Ladak A, et al. Brief electrical stimulation improves nerve regeneration after delayed repair in Sprague Dawley rats. Exp Neurol 2015;269:142-53. [Crossref] [PubMed]
- Yang IH, Gary D, Malone M, et al. Axon Myelination and Electrical Stimulation in a Microfluidic, Compartmentalized Cell Culture Platform. NeuroMolecular Med 2012;14:112-8. [Crossref] [PubMed]
- Feinberg AW, Alford PW, Jin H, et al. Controlling the contractile strength of engineered cardiac muscle by hierarchal tissue architecture. Biomaterials 2012;33:5732-41. [Crossref] [PubMed]
- Tandon N, Cannizzaro C, Chao P-HG, et al. Electrical stimulation systems for cardiac tissue engineering. Nat Protoc 2009;4:155-73. [Crossref] [PubMed]
- Pietronave S, Zamperone A, Oltolina F, et al. Monophasic and Biphasic Electrical Stimulation Induces a Precardiac Differentiation in Progenitor Cells Isolated from Human Heart. Stem Cells Dev 2014;23:888-98. [Crossref] [PubMed]
- Hamnström J. Design of a Smartphone App for Control of Bioreactors Used for Cell Cultivation. Available online: https://uu.diva-portal.org/smash/get/diva2:516669/FULLTEXT01.pdf
- Olivo J, Foglia L, Casulli MA, et al. Glucose and lactate monitoring in cell cultures with a wireless android interface. In: 2014 IEEE Biomedical Circuits and Systems Conference (BioCAS) Proceedings. Piscataway Township: IEEE, 2014;400-3.
- Stradolini F, Riario S, Boero C, et al. Wireless Monitoring of Endogenous and Exogenous Biomolecules on an Android Interface. IEEE Sens J 2016;16:3163-70. [Crossref]
- Zhang YS, Busignani F, Ribas J, et al. Google Glass-Directed Monitoring and Control of Microfluidic Biosensors and Actuators. Sci Rep 2016;6:22237. [Crossref] [PubMed]
- Stradolini F, Lavalle E, De Micheli G, et al. Paradigm-shifting players for iot: Smart-watches for intensive care monitoring. Berlin: Springer, LNICST, 2017;19:71-8.
- Riahi R, Shaegh SAM, Ghaderi M, et al. Automated microfluidic platform of bead-based electrochemical immunosensor integrated with bioreactor for continual monitoring of cell secreted biomarkers. Sci Rep 2016;6:24598. [Crossref] [PubMed]
- Shin SR, Zhang YS, Kim DJ, et al. Aptamer-Based Microfluidic Electrochemical Biosensor for Monitoring Cell-Secreted Trace Cardiac Biomarkers. Anal Chem 2016;88:10019-27. [Crossref] [PubMed]
- Veal EA, Day AM, Morgan BA. Hydrogen Peroxide Sensing and Signaling. Mol Cell 2007;26:1-14. [Crossref] [PubMed]
- Rhee SG. H2O2, a necessary evil for cell signaling. Science 2006;312:1882-3. [Crossref] [PubMed]
- Ge S, Liu W, Liu H, et al. Colorimetric detection of the flux of hydrogen peroxide released from living cells based on the high peroxidase-like catalytic performance of porous PtPd nanorods. Biosens Bioelectron 2015;71:456-62. [Crossref] [PubMed]
- Zhang LN, Deng HH, Lin FL, et al. In situ growth of porous platinum nanoparticles on graphene oxide for colorimetric detection of cancer cells. Anal Chem 2014;86:2711-8. [Crossref] [PubMed]
- Tsaplev YB. Chemiluminescence determination of hydrogen peroxide. J Anal Chem 2012;67:506-14. [Crossref]
- Gao X, Ding C, Zhu A, et al. Carbon-dot-based ratiometric fluorescent probe for imaging and biosensing of superoxide anion in live cells. Anal Chem 2014;86:7071-8. [Crossref] [PubMed]
- Zhuang M, Ding C, Zhu A, et al. Ratiometric fluorescence probe for monitoring hydroxyl radical in live cells based on gold nanoclusters. Anal Chem 2014;86:1829-36. [Crossref] [PubMed]
- Wen F, Dong Y, Feng L, et al. Horseradish peroxidase functionalized fluorescent gold nanoclusters for hydrogen peroxide sensing. Anal Chem 2011;83:1193-6. [Crossref] [PubMed]
- Wang T, Zhu H, Zhuo J, et al. Biosensor based on ultrasmall MoS2 nanoparticles for electrochemical detection of H2O2 released by cells at the nanomolar level. Anal Chem 2013;85:10289-95. [Crossref] [PubMed]
- Zhang Y, Bai X, Wang X, et al. Highly Sensitive Graphene-Pt Nanocomposites Amperometric Biosensor and Its Application in Living Cell H2O2 Detection. Anal Chem 2014;86:9459-65. [Crossref] [PubMed]
- Liu F, Bing T, Shangguan D, et al. Ratiometric Fluorescent Biosensing of Hydrogen Peroxide and Hydroxyl Radical in Living Cells with Lysozyme-Silver Nanoclusters: Lysozyme as Stabilizing Ligand and Fluorescence Signal Unit. Anal Chem 2016;88:10631-8. [Crossref] [PubMed]
- Chen X, Wu G, Cai Z, et al. Advances in enzyme-free electrochemical sensors for hydrogen peroxide, glucose, and uric acid. Microchimica Acta 2014;181:689-705. [Crossref]
- Chen S, Yuan R, Chai Y, et al. Electrochemical sensing of hydrogen peroxide using metal nanoparticles: A review. Microchim Acta 2013;180:15-32. [Crossref]
- Tiwari JN, Vij V, Kemp KC, et al. Engineered carbon-nanomaterial-based electrochemical sensors for biomolecules. ACS Nano 2016;10:46-80. [Crossref] [PubMed]
- Zhao J, Yan Y, Zhu L, et al. An amperometric biosensor for the detection of hydrogen peroxide released from human breast cancer cells. Biosens Bioelectron 2013;41:815-9. [Crossref] [PubMed]
- Maji SK, Sreejith S, Mandal AK, et al. Immobilizing gold nanoparticles in mesoporous silica covered reduced graphene oxide: a hybrid material for cancer cell detection through hydrogen peroxide sensing. ACS applied materials & interfaces 2014;6:13648-56. [Crossref] [PubMed]
- Ju J, Chen W. In situ growth of surfactant-free gold nanoparticles on nitrogen-doped graphene quantum dots for electrochemical detection of hydrogen peroxide in biological environments. Anal Chem 2015;87:1903-10. [Crossref] [PubMed]
- Liu J, Bo X, Zhao Z, et al. Highly exposed Pt nanoparticles supported on porous graphene for electrochemical detection of hydrogen peroxide in living cells. Biosens Bioelectron 2015;74:71-7. [Crossref] [PubMed]
- Xiao F, Li Y, Zan X, et al. Growth of metal-metal oxide nanostructures on freestanding graphene paper for flexible biosensors. Adv Funct Mater 2012;22:2487-94. [Crossref]
- Mathupala SP, Colen CB, Parajuli P, et al. Lactate and malignant tumors: A therapeutic target at the end stage of glycolysis. Journal of Bioenergetics and Biomembranes 2007;39:73-7. [Crossref] [PubMed]
- Mongersun A, Smeenk I, Pratx G, et al. Droplet Microfluidic Platform for the Determination of Single-Cell Lactate Release. Anal Chem 2016;88:3257-63. [Crossref] [PubMed]
- Simmons AD, Williams C, Degoix A, et al. Sensing metabolites for the monitoring of tissue engineered construct cellularity in perfusion bioreactors. Biosens Bioelectron 2017;90:443-9. [Crossref] [PubMed]
- Zheng XT, Yang HB, Li CM. Optical detection of single cell lactate release for cancer metabolic analysis. Anal Chem 2010;82:5082-7. [Crossref] [PubMed]
- Bavli D, Prill S, Ezra E, et al. Real-time monitoring of metabolic function in liver-on-chip microdevices tracks the dynamics of mitochondrial dysfunction. Proc Natl Acad Sci U S A 2016;113:E2231-40. [Crossref] [PubMed]
- Ges IA, Baudenbacher F. Enzyme-coated microelectrodes to monitor lactate production in a nanoliter microfluidic cell culture device. Biosens Bioelectron 2010;26:828-33. [Crossref] [PubMed]
- Lin Y, Yu P, Hao J, et al. Continuous and simultaneous electrochemical measurements of glucose, lactate, and ascorbate in rat brain following brain ischemia. Anal Chem 2014;86:3895-901. [Crossref] [PubMed]
- Cheng W, Klauke N, Sedgwick H, et al. Metabolic monitoring of the electrically stimulated single heart cell within a microfluidic platform. Lab Chip 2006;6:1424-31. [Crossref] [PubMed]
- Pemberton RM, Cox T, Tuffin R, et al. Fabrication and evaluation of a micro(bio)sensor array chip for multiple parallel measurements of important cell biomarkers. Sensors (Basel) 2014;14:20519-32. [Crossref] [PubMed]
- Weltin A, Slotwinski K, Kieninger J, et al. Cell culture monitoring for drug screening and cancer research: a transparent, microfluidic, multi-sensor microsystem. Lab Chip 2014;14:138-46. [Crossref] [PubMed]
- Weltin A, Hammer S, Noor F, et al. Accessing 3D microtissue metabolism: Lactate and oxygen monitoring in hepatocyte spheroids. Biosens Bioelectron 2017;87:941-8. [Crossref] [PubMed]
- Prill S, Jaeger MS, Duschl C. Long-term microfluidic glucose and lactate monitoring in hepatic cell culture. Biomicrofluidics 2014;8:034102 [Crossref] [PubMed]
- Frey O, Misun PM, Rothe J, et al. Real-time in-situ lactate monitoring in 3D multi-cellular spheroid cultures by using enzyme-based biosensors in hanging drop networks. In: Procedia Engineering. Amsterdam: Elsevier, 2014;96-9.
- Nichols SP, Koh A, Storm WL, et al. Biocompatible materials for continuous glucose monitoring devices. Chemical Reviews 2013;113:2528-49. [Crossref] [PubMed]
- Stenken JA, Poschenrieder AJ. Bioanalytical chemistry of cytokines - A review. Analytica Chimica Acta 2015;853:95-115. [Crossref] [PubMed]
- Liu G, Qi M, Hutchinson MR, et al. Recent advances in cytokine detection by immunosensing. Biosensors and Bioelectronics 2016;79:810-21. [Crossref] [PubMed]
- Matharu Z, Patel D, Gao Y, et al. Detecting transforming growth factor-β release from liver cells using an aptasensor integrated with microfluidics. Anal Chem 2014;86:8865-72. [Crossref] [PubMed]
- Ma C, Fan R, Ahmad H, et al. A clinical microchip for evaluation of single immune cells reveals high functional heterogeneity in phenotypically similar T cells. Nat Med 2011;17:738-43. [Crossref] [PubMed]
- Yu ZT, Guan H, Cheung MK, et al. Rapid, automated, parallel quantitative immunoassays using highly integrated microfluidics and AlphaLISA. Sci Rep 2015;5:11339. [Crossref] [PubMed]
- Chokkalingam V, Tel J, Wimmers F, et al. Probing cellular heterogeneity in cytokine-secreting immune cells using droplet-based microfluidics. Lab Chip 2013;13:4740. [Crossref] [PubMed]
- Liu Y, Liu Y, Matharu Z, et al. Detecting multiple cell-secreted cytokines from the same aptamer-functionalized electrode. Biosens Bioelectron 2015;64:43-50. [Crossref] [PubMed]
- Zhou Q, Kwa T, Gao Y, et al. On-chip regeneration of aptasensors for monitoring cell secretion. Lab Chip 2014;14:276-9. [Crossref] [PubMed]
- Zhou Q, Patel D, Kwa T, et al. Liver injury-on-a-chip: microfluidic co-cultures with integrated biosensors for monitoring liver cell signaling during injury. Lab Chip 2015;15:4467-78. [Crossref] [PubMed]
- Son KJ, Gheibi P, Stybayeva G, et al. Detecting cell-secreted growth factors in microfluidic devices using bead-based biosensors. Microsystems & Nanoengineering 2017;3:17025. [Crossref] [PubMed]
- Shin SR, Kilic T, Zhang YS, et al. Label-Free and Regenerative Electrochemical Microfluidic Biosensors for Continual Monitoring of Cell Secretomes. Adv Sci (Weinh) 2017;4:1600522 [Crossref] [PubMed]
- Murphy SV, Anthony A. Regenerative Medicine Technology: On-a-Chip Applications for Disease Modeling, Drug Discovery and Personalised Medicine. Boca Raton: CRC Press, 2017.
- Wikswo JP, Block FE, Cliffel DE, et al. Engineering challenges for instrumenting and controlling integrated organ-on-chip systems. IEEE Transactions on Biomedical Engineering 2013;60:682-90. [Crossref] [PubMed]
- Solaini G, Baracca A, Lenaz G, et al. Hypoxia and mitochondrial oxidative metabolism. Biochim Biophys Acta 2010;1797:1171-7. [Crossref] [PubMed]
- Bavli D, Prill S, Ezra E, et al. Real-time monitoring of metabolic function in liver-on-chip microdevices tracks the dynamics of mitochondrial dysfunction. Proc Natl Acad Sci U S A 2016;113:E2231-40. [Crossref] [PubMed]
- Oomen PE, Skolimowski MD, Verpoorte E. Implementing oxygen control in chip-based cell and tissue culture systems. Lab Chip 2016;16:3394-414. [Crossref] [PubMed]
- Nastuk WL. Electrophysiological Methods: Physical Techniques in Biological Research. Amsterdam: Elsevier, 2013.
- Suzuki H, Hirakawa T, Watanabe I, et al. Determination of blood pO2 using a micromachined Clark-type oxygen electrode. Anal Chim Acta 2001;431:249-59. [Crossref]
- Wu CC, Yasukawa T, Shiku H, et al. Fabrication of miniature Clark oxygen sensor integrated with microstructure. Sensors Actuators B 2005;110:342-9. [Crossref]
- Parizi KB, Xu X, Pal A, et al. ISFET pH Sensitivity: Counter-Ions Play a Key Role. Sci Rep 2017;7:41305. [Crossref] [PubMed]
- Krommenhoek EE, Van Leeuwen M, Gardeniers H, V, et al. Lab-scale fermentation tests of microchip with integrated electrochemical sensors for pH, temperature, dissolved oxygen and viable biomass concentration. Biotechnol Bioeng 2008;99:884-92. [Crossref] [PubMed]
- Allen JR. pH Electrodes, Ion-Selective Electrodes, and Oxygen Sensors: Electrochemical Sensors Used in the Medical Field. Laboratory Medicine 2003;34:544-7. [Crossref]
- Brennan MD, Rexius-Hall ML, Elgass LJ, et al. Oxygen control with microfluidics. Lab Chip 2014;14:4305-18. [Crossref] [PubMed]
- Shoda M, Ishikawa Y. Carbon dioxide sensor for fermentation systems. Biotechnol Bioeng 1981;23:461-6. [Crossref]
- Sud D, Mehta G, Mehta K, et al. Optical imaging in microfluidic bioreactors enables oxygen monitoring for continuous cell culture. J Biomed Opt 2006;11:050504 [Crossref] [PubMed]
- Lee SK, Okura I. Photoluminescent determination of oxygen using metalloporphyrin-polymer sensing systems. Spectrochim Acta Part A 1998;54:91-100. [Crossref]
- Vanderkooi JM, Maniara G, Green TJ, et al. An optical method for measurement of dioxygen concentration based upon quenching of phosphorescence. J Biol Chem 1987;262:5476-82. [PubMed]
- Prill S, Bavli D, Levy G, et al. Real-time monitoring of oxygen uptake in hepatic bioreactor shows CYP450-independent mitochondrial toxicity of acetaminophen and amiodarone. Arch Toxicol 2016;90:1181-91. [Crossref] [PubMed]
- Mehta G, Mehta K, Sud D, Song JW, Bersano-Begey T, Futai N, et al. Quantitative measurement and control of oxygen levels in microfluidic poly(dimethylsiloxane) bioreactors during cell culture. Biomed Microdevices 2007;9:123-34. [Crossref] [PubMed]
- Park EJ, Reid KR, Tang W, et al. Ratiometric fiber optic sensors for the detection of inter- and intra-cellular dissolved oxygen. J Mater Chem 2005;15:2913. [Crossref]
- Wirnsberger G, Scott BJ, Stucky GD, et al. pH Sensing with mesoporous thin films. Chem Commun 2001;405:119-20. [Crossref]
- Hiruta Y, Ando Y, Citterio D, et al. A fast-response pH optode based on a fluoroionophore immobilized to a mesoporous silica thin film. Anal Sci 2010;26:297-301. [Crossref] [PubMed]
- Bergveld P. Development of an Ion-Sensitive Solid-State Device for Neurophysiological Measurements. IEEE Trans Biomed Eng 1970;17:70-1. [Crossref] [PubMed]
-
ISFET, Theory and Practice - Wu L, Gao X, Brown RC, et al. Dual role of the TRPV4 channel as a sensor of flow and osmolality in renal epithelial cells. Am J Physiol Renal Physiol 2007;293:F1699-713. [Crossref] [PubMed]
- Lehmann M, Baumann W, Brischwein M, et al. Simultaneous measurement of cellular respiration and acidification with a single CMOS ISFET. Biosens Bioelectron 2001;16:195-203. [Crossref] [PubMed]
- Young EW, Beebe DJ. Fundamentals of microfluidic cell culture in controlled microenvironments. Chem Soc Rev 2010;39:1036-48. [Crossref] [PubMed]
- Mattes R, Root D, Sugui MA, et al. Real-time bioreactor monitoring of osmolality and pH using near-infrared spectroscopy. Bioprocess Int 2009;7:44-50.
- Zhang YS, Ribas J, Nadhman A, et al. A Cost-Effective Fluorescence Mini-Microscope with Adjustable Magnifications for Biomedical Applications. Lab Chip 2015;15:3661-9. [Crossref] [PubMed]
- Joeris K, Frerichs J-G, Konstantinov K, et al. In-situ microscopy: Online process monitoring of mammalian cell cultures. Cytotechnology 2002;38:129-34. [Crossref] [PubMed]
- Stephens JS, Cooper JA, Phelan FR, et al. Perfusion flow bioreactor for 3D in situ imaging: Investigating cell/biomaterials interactions. Biotechnol Bioeng 2007;97:952-61. [Crossref] [PubMed]
- Psaltis D, Quake SR, Yang C. Developing optofluidic technology through the fusion of microfluidics and optics. Nature 2006;442:381-6. [Crossref] [PubMed]
- Xu Y, Xie X, Duan Y, et al. A review of impedance measurements of whole cells. Biosens Bioelectron 2016;77:824-36. [Crossref] [PubMed]
- Wang H, Sobahi N, Han A. Impedance spectroscopy-based cell/particle position detection in microfluidic systems. Lab Chip 2017;17:1264-9. [Crossref] [PubMed]
- Danquah MK, Mahato RI. Emerging Trends in Cell and Gene Therapy. New York: Humana Press, 2013.
- Kumar S, Kumar S, Ali MA, et al. Microfluidic-integrated biosensors: prospects for point-of-care diagnostics. Biotechnol J 2013;8:1267-79. [Crossref] [PubMed]
- Wang Y, Chen Q, Zeng X. Potentiometric biosensor for studying hydroquinone cytotoxicity in vitro. Biosens Bioelectron 2010;25:1356-62. [Crossref] [PubMed]
- Asphahani F, Zhang M. Cellular impedance biosensors for drug screening and toxin detection. Analyst 2007;132:835-41. [Crossref] [PubMed]
- Heileman K, Daoud J, Tabrizian M. Dielectric spectroscopy as a viable biosensing tool for cell and tissue characterization and analysis. Biosensors and Bioelectronics 2013;49:348-59. [Crossref] [PubMed]
- Holmes D, Pettigrew D, Reccius CH, et al. Leukocyte analysis and differentiation using high speed microfluidic single cell impedance cytometry. Lab Chip 2009;9:2881. [Crossref] [PubMed]
- Arndt S, Seebach J, Psathaki K, et al. Bioelectrical impedance assay to monitor changes in cell shape during apoptosis. Biosens Bioelectron 2004;19:583-94. [Crossref] [PubMed]
- Asphahani F, Wang K, Thein M, et al. Single-cell bioelectrical impedance platform for monitoring cellular response to drug treatment. Phys Biol 2011;8:015006 [Crossref] [PubMed]
- Thein M, Asphahani F, Cheng A, et al. Biosensors and Bioelectronics Response characteristics of single-cell impedance sensors employed with surface-modified microelectrodes. Biosens Bioelectron 2010;25:1963-9. [Crossref] [PubMed]
- Senez V, Lennon E, Ostrovidov S, et al. Integrated 3-D Silicon Electrodes for Electrochemical Sensing in Microfluidic Environments : Application to Single-Cell Characterization. IEEE Sens J 2008;8:548-57. [Crossref]
- Asami K. Simulation for the dielectric images of single biological cells obtained using a scanning dielectric microscope. Journal of Physics D: Applied Physics 2008;41:085501 [Crossref]
- Luong JH, Habibi-Rezaei M, Meghrous J, et al. Monitoring Motility, Spreading, and Mortality of Adherent Insect Cells Using an Impedance Sensor. Anal Chem 2001;73:1844-8. [Crossref] [PubMed]
- Yoo EH, Lee SY. Glucose biosensors: an overview of use in clinical practice. Sensors (Basel) 2010;10:4558-76. [Crossref] [PubMed]
- Turner AP. Biosensors: sense and sensibility. Chem Soc Rev 2013;42:3184-96. [Crossref] [PubMed]
- Scognamiglio V. Biosensors and Bioelectronics Nanotechnology in glucose monitoring : Advances and challenges in the last 10 years. Biosens Bioelectron 2013;47:12-25. [Crossref] [PubMed]
- Jeffery CJ. Engineering periplasmic ligand binding proteins as glucose nanosensors. Nano Rev 2011;2. [PubMed]
- Scognamiglio V, Scirè A, Aurilia V, et al. A strategic fluorescence labeling of D-galactose/D-glucose-binding protein from Escherichia coli helps to shed light on the protein structural stability and dynamics. J Proteome Res 2007;6:4119-26. [Crossref] [PubMed]
- Ghenim L, Kaji H, Hoshino Y, et al. Monitoring impedance changes associated with motility and mitosis of a single cell. Lab Chip 2010;10:2546. [Crossref] [PubMed]
- Bagnaninchi PO, Drummond N. Real-time label-free monitoring of adipose-derived stem cell differentiation with electric cell-substrate impedance sensing. Proc Natl Acad Sci 2011;108:6462-7. [Crossref] [PubMed]
- Angstmann M, Brinkmann I, Bieback K, et al. Monitoring human mesenchymal stromal cell differentiation by electrochemical impedance sensing. Cytotherapy 2011;13:1074-89. [Crossref] [PubMed]
- Cho S, Gorjup E, Thielecke H. Chip-based time-continuous monitoring of toxic effects on stem cell differentiation. Ann Anat 2009;191:145-52. [Crossref] [PubMed]
- Xie L, Yan X, Du Y. Biosensors and Bioelectronics An aptamer based wall-less LSPR array chip for label-free and high throughput detection of biomolecules. Biosens Bioelectron 2014;53:58-64. [Crossref] [PubMed]
- Hu N, Wu C, Ha D, et al. Biosensors and Bioelectronics A novel microphysiometer based on high sensitivity LAPS and microfluidic system for cellular metabolism study and rapid drug screening. Biosens Bioelectron 2013;40:167-73. [Crossref] [PubMed]
- Bhagat AA, Bow H, Hou HW, et al. Microfluidics for cell separation. Med Biol Eng Comput 2010;48:999-1014. [Crossref] [PubMed]
- Shields CW, Reyes D, López GP. Lab on a Chip in the separation of cells from debulking to rare. Lab Chip 2015;15:1230-49. [Crossref] [PubMed]
- Tothill IE. Biosensors for cancer markers diagnosis. Semin Cell Dev Biol 2009;20:55-62. [Crossref] [PubMed]
- TermehYousefi A. Bagheri S, Adib N. Integration of biosensors based on microfluidic : a review. Sensor Review 2015;35:190-9. [Crossref]
- Bhagat AA, Kuntaegowdanahalli SS, Papautsky I. Inertial microfluidics for continuous particle filtration and extraction. Microfluid Nanofluidics 2009;7:217-26. [Crossref]
- Inglis DW, Davis JA, Austin RH, et al. Critical particle size for fractionation by deterministic lateral displacement. Lab Chip 2006;6:655. [Crossref] [PubMed]
- Yamada M, Kano K, Tsuda Y, et al. Microfluidic devices for size-dependent separation of liver cells. Biomed Microdevices 2007;9:637-45. [Crossref] [PubMed]
- Choi S, Karp JM, Karnik R. Cell sorting by deterministic cell rolling. Lab Chip 2012;12:1427. [Crossref] [PubMed]
- Mazutis L, Gilbert J, Ung WL, et al. Single-cell analysis and sorting using droplet-based microfluidics. Nat Protoc 2013;8:870-91. [Crossref] [PubMed]
- Fu AY, Spence C, Scherer A, Arnold FH, Quake SR. A microfabricated fluorescence-activated cell sorter. Nat Biotechnol 1999;17:1109-11. [Crossref] [PubMed]
- Wang X, Chen S, Kong M, et al. Enhanced cell sorting and manipulation with combined optical tweezer and microfluidic chip technologies. Lab Chip 2011;11:3656. [Crossref] [PubMed]
- Shields CW, Reyes CD, López GP. Microfluidic cell sorting: a review of the advances in the separation of cells from debulking to rare cell isolation. Lab Chip 2015;15:1230-49. [Crossref] [PubMed]
- Ozkumur E, Shah AM, Ciciliano JC, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med 2013;5:179ra47 [Crossref] [PubMed]
- Shields CW, Johnson LM, Gao L, et al. Elastomeric negative acoustic contrast particles for capture, acoustophoretic transport, and confinement of cells in microfluidic systems. Langmuir 2014;30:3923-7. [Crossref] [PubMed]
- Kim U, Soh HT. Simultaneous sorting of multiple bacterial targets using integrated Dielectrophoretic–Magnetic Activated Cell Sorter. Lab Chip 2009;9:2313. [Crossref] [PubMed]
- Tzouvadaki I, Jolly P, Lu X, et al. Label-free ultrasensitive memristive aptasensor. Nano Lett 2016;16:4472-6. [Crossref] [PubMed]
Cite this article as: Kilic T, Navaee F, Stradolini F, Renaud P, Carrara S. Organs-on-chip monitoring: sensors and other strategies. Microphysiol Syst 2018;2:5.