
Gut-microbiota-on-a-chip: an enabling field for physiological research
Introduction
The stability and assembled function of our intestinal microbiota is key to determining (and assessing) health (1-4) and disease (5-7). Alteration in the normal human microbiota, referred to as dysbiosis (5,8,9), has been associated with many diseases (5) and syndromes, such as diabetes (10-12), autoimmune diseases (13-15), allergies (8,16,17), inflammatory bowel disease (9,18,19), obesity (20-22), metabolic syndrome (23), hepatic inflammatory disease (24), and some forms of cancer (3,7,25,26). A growing body of evidence supports the role of the gut microbiota (and its metabolites) in the regulation of blood pressure (27), and the interplay between our immune system and our microbiota (1,8,13). Not surprisingly, the study of human microbiota (and the microbiome) has accelerated (4,15,28,29) during the last decade. The cumulative technical knowledge on the origin, dynamics, and importance of human gut microbiota has enabled significant advances in biomedicine and nutrition.
However, the study of the host-microbiota interaction in vivo is limited by the inaccessibility of the digestive tract. Therefore, the study of the interplay between the human host and its microbiota has been mainly limited to the analysis of human fecal samples (30,31) and studies in animal models (mainly mice) (32,33). Nevertheless, these platforms have inherent limitations. For example, experimentation with human volunteers is expensive and time-consuming. Moreover, in human subjects, the effect of a specific experimental variable is difficult to assess independently. Research using animal models is also expensive, and not always scalable or fully significant to humans (34,35). Research with animal models or with humans must also adhere to strict ethical and legal frameworks that understandably limit scope and speed.
These limitations can be overcome by the use of in vitro systems based on two-dimensional (2D) cell culture methodologies. These systems have been used to study basic features of the complex processes that take place in the human intestine, such as the absorption of pharmaceutical compounds and nutrients and the dynamics of parasite infection, among others (36,37). However, experimental evidence has shown that 2D cell culture systems fail to reproduce some relevant aspects of the complex architecture and dynamics (i.e., morphophysiology) of the human intestine (36-39). It is anticipated that, the availability of more physiologically relevant in vitro platforms will further propel research on the interplay between microbiota and host cells. These platforms should recapitulate some of the key aspects of the dynamics of the human gut, a highly complex environment.
One active front of research is the development of gut-on-chip models that are capable of emulating some relevant features and dynamic behaviors of the human intestinal tract. This approach is challenging due to the complexity of both the architecture and function of the human gut and its microbiota. However, gut-on-chip systems have the potential to provide answers that cannot be obtained using conventional experimental platforms. The gut-on-chip platforms enable a fine control of culture conditions and could lead to the implementation of high-throughput analytical techniques.
The aim of this paper is to review the state of the art and to survey the advances in the gut-on-chip technologies that directly relate to improving our understanding of the complex interplay between gut microbiota and human physiology. We start by briefly describing the composition, physiological relevance, and basic dynamics of human microbiota. We then comment on the architecture of the human intestine and the general approaches used to select biomaterials and scaffolds to recapitulate some aspects of the interaction between the physical environment of the intestine, the key bacterial actors of the human microbiota, and human intestinal cells. We also review, in detail, the relevant and recent information related to the conceptual design, fabrication, and applications of gut-on-chip systems, focusing on the study of the interplay between the human host and the human microbiota. In particular, we discuss a set of pioneering examples of gut-on-chip systems that aim to emulate some relevant functional features and events related to the interactions between human gut tissues and intestinal microbiota. We describe these from simple to more complex in terms of system architecture and dynamics and the degree of interaction between microbiota (one or more types of bacteria) and human cells (i.e., intestinal epithelial cells, immune cytokines). We then comment on the use of microfabrication techniques (particularly 3D printing and bioprinting) and stem cell strategies, two powerful enablers of next-generation gut microbiota-on-chip platforms. Finally, we briefly refer to some of the opportunities and challenges that we foresee regarding the use of gut microbiota-on-chip systems to address relevant and pressing problems related to human health.
The relevance and complexity of human microbiota in a nutshell
Although the estimates vary widely, a recent report concludes that a 70-kg reference human is inhabited by approximately 1013 symbiotic bacteria (roughly 0.2 kg in dry weight) (40) that reside primarily in the intestine. The number of human cells in this average individual would be within the same order of magnitude, if erythrocytes (non-nucleated cells) are included in the calculation (Figure 1A). Therefore, a ratio of 1:1 for bacteria:human cells appears to be a more realistic figure than the frequently referred (42,43) [and recently questioned (40,44)] 10:1 ratio (note that this ratio holds true if erythrocytes are not counted). These revised numbers justify the hypothesis that our microbiota is at least as important as our own cells, in the context of human physiology and health.
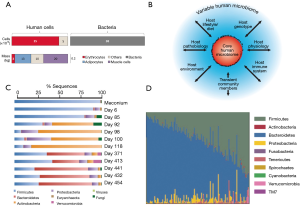
At the level of the intestine, the ratio between bacteria and intestinal cells becomes impressively high. The trillions of microbes (45-47) that inhabit the human intestine form a complex ecological community (4) whose collective genome contribution, the “microbiome”, contains at least two orders of magnitude more genes that our own human genome (47). These remarkably high numbers of bacteria in the human gut have physiological purposes (4,47). For one, microbiota greatly enriches our food processing capabilities, providing us with a metabolic arsenal that facilitates the digestion of materials that we are unable to process with our own enzymatic machinery (47).
The composition of human gut microbiota is highly complex and has only been partially elucidated (4,42,45,48). Several different studies have identified a common core of bacteria in the human intestine (4,31,42,47,49,50) while it harbors a stunningly diverse array of microbial species [100–1,000 species (51)]. Although very diverse in terms of species and strains, more than 90% of the phylogenetic types (phylotypes) found in the human intestine belong to just two of the 70 known divisions of bacteria, namely, the Bacteroidetes and the Firmicutes; the remaining phylotypes are distributed among eight other divisions (52-54).
As previously referred, current evidence confirms that the composition of gut microbiota and its degree of diversity play a crucial role in human health and disease (5,6). Consequently, understanding the cross-talk between the gut environment and the dynamics of the microbiota, as well as the evolution of our microbiota through life, has received particular attention in the last decade. A brief summary of our current knowledge on these particular topics follows.
The gut microbiota appears to have evolved with humans (49,50,54) in an intimate symbiosis that has occurred over many millennia. Nevertheless, the gut microbiome is not significantly associated with genetic ancestry, and the host genetics play only a minor role in determining the ultimate microbiome composition (55). By contrast, the composition of the microbiome of any single individual (4,20,50) is significantly influenced by multiple environmental factors (13,29,56), including diet and exposure to food additives (57) and drugs (i.e., antibiotics) (58,59). Over 20% of the inter-person microbiome variability has been associated with environmental factors (Figure 1B) (55). Indeed, significant variation is observed in the core composition of the microbiota in different geographical and cultural settings, and a similarity in the gut microbiota among family members has been documented (60). Significant similarities also exist in the compositions of the microbiomes of genetically unrelated individuals who share a household (55).
Within an individual, the microbiota composition varies across age (2,60,61). A share of microbiota “memory” is present at birth, as skin or vaginal microbiota are transferred from mother to child during delivery (62) and breastfeeding (63). Throughout early development, a significant fluctuation in microbiota has been documented (Figure 1C) in any given individual (6,41); gut microbiota evolves in time from a community dominated by the bacteria acquired during birth, which help the infant to assimilate certain nutrients and boost its immune system, to a relatively constant core composition in the healthy adult dominated by Bacteroidetes and Firmicutes (2,53). Variations in microbiota among healthy individuals in a given population appear to be much higher than the variation within a healthy individual during adulthood (Figure 1D) (4). However, if a healthy subject is exposed to out-of-the-ordinary events, such as a drastic change in diet, an acute infection, or antibiotic administration, the composition of the microbiota could be severely disturbed, but it eventually and resiliently returns to the previous-to-disturbance status (4,45).
Cells, biomaterials, and scaffolds useful to study microbiota-host interactions
Several reaction/culture systems (human-gut-simulators) are in use to culture the human microbiota and recreate its composition and biochemical activity in different sections of the gastrointestinal tract (56,64,65). These systems, conventionally composed of a series of reactors in the scale of hundreds of milliliters of volume, are frequently used in the context of absorption/nutrition studies. However, most previously reported gut-on-chip devices do not attempt to recreate the highly complex composition and dynamics of the human gut microbiota; rather, they aim to recapitulate relevant pieces of it, and they do provide valuable information. Indeed, most of the reported experiments on microbiota-on-chip systems have considered one or two bacterial strains, with only a few reported results with bacterial species. The most commonly used bacterial strain in gut-on-chip systems is Escherichia coli (66), which is frequently used as a commensal model (67-69). In some reports, pathogenic entero-invasive E. coli strains have been used to model invasion or infection by pathogenic bacteria into the commensal bacterial biofilm or the host intestinal endothelium (67,68). Other pathogenic bacteria used in gut-on-chips include Staphylococcus aureus, Pseudomonas aeruginosa, and Salmonella typhimurium (69,70). Some lactic acid bacteria have been used to model the presence or the biochemical contribution of beneficial bacteria (probiotic bacteria) in human microbiota (69,71,72).
The complexity of the composition and dynamics of the human gut microbiota is only one of several important aspects to consider in the design of relevant microbiota-on-chip systems. One challenge is to develop intestinal scaffolds that are capable of reproducing the complex multi-scale and multi-layered architecture, as well as surface topography and function, of the human intestine. Figure 2A shows a simplified graphical representation of the architecture of the intestinal tissue. The intestine has folds (better known as circular folds) that increase the surface area available for optimized absorption of nutrients. The epithelium of the human small intestinal possesses a distinct crypt-villus architecture (31). The villi are finger- or tongue-like microstructures (depending on the portion of the intestine) that are formed by epithelial cells and protrude into the intestinal lumen. With an average height and base length of 550 and 160 μm, respectively (77), the villi are the minimum functional unit of intestinal tissue. The valleys between the villi are known as crypts. The crypt-villus axes exhibit a characteristic tissue polarity, in which proliferative cells reside inside crypts whereas mainly differentiated cells form the villi (78,79).
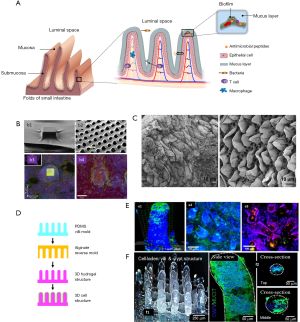
Another important player in the intestinal dynamical equilibrium between microbiota and human cells is the intestinal mucus (80). The intestinal epithelium is covered with a mucus layer approximately 100-µm in thickness (81) that is secreted by the goblet cells (80). The mucus layer, long underappreciated, limits the direct contact between bacteria and human cells and slows down bacterial penetration into the epithelial monolayer, while concentrating the anti-bacterial peptides produced by the crypt Paneth cells. Without mucus, the anti-bacterial peptides would be quickly diluted into the luminal content. Therefore, the mucus helps to establish an antibacterial chemical gradient.
In recent years, several researchers have engineered scaffolds that resemble specific features of the architecture, topography, and functionality of the human intestine (4,69,70,76,82). For example, the epithelial layer in the intestine plays a fundamental role as a permeable membrane that allows the transport of materials from the lumen to the blood stream. Several approaches have been used to specifically mimic this permeable barrier function. For instance, Esch et al. described the fabrication of permeable polymeric membranes of tunable 3D shapes that could be integrated into microfluidic systems to emulate the functions of physiological membranes (73). These authors developed a simple method based on the partial exposure of photoresist (SU-8) on flat silicon substrates to produce flat membranes with pores of 0.5 to 4.0 mm in diameter. Alternatively, thin layers of SU-8 were dispensed and air-dried over silicon pillars of specific 3D shapes (Figure 2B) to fabricate porous membranes that resembled the villus intestinal architecture and served as a scaffold for the culture of Caco-2 cells. These microfabricated membranes could be used to integrate barrier tissues (e.g., the gastrointestinal tract epithelium, the lung epithelium, or other barrier tissues) within multi-organ “human-on-a-chip” devices.
Alternatively, intestinal tissues might be decellularized and used for engineering the gut microstructure. In fact, the use of decellularized gut tissues has been suggested (74), but not yet implemented, in the development of microbiota-gut-on-chip platforms. Several simple protocols have been reported for decellularization of intestinal tissue, mainly for tissue engineering applications (74,83,84). For instance, Totonelli et al. used decellularized rat intestinal tissue as a scaffold that preserved the architecture of the intestinal (duodenal) tongue-shaped villi (Figure 2C) (74). Briefly, both the intestinal lumen and the vascular tree were perfused with continuous fluid delivery using a peristaltic pump at 0.6 mL/h. Each detergent cycle consisted of successive and slow flowing of (I) deionized water (resistivity 18.2 MΩ/cm) at 4 °C for 24 h, (II) 4% sodium deoxycholate at room temperature for 4 h, and (III) 2,000-kU DNase-I in 1 M NaCl at a residence time of 3 hours (each solution). After each treatment cycle, the constructs were preserved at 4 °C, in phosphate buffer saline (PBS) containing 5% of antibiotic-antimycotic solution. Subsequent histological examination and scanning electron microscopy (SEM) and transmission electron microscopy (TEM) analyses confirmed the removal of cellular elements while preserving the native architecture of the tissue. Moreover, the decellularized tissue exhibited similar mechanical stiffness to that of the native tissue, adequate cell adhesion (as evaluated by culturing amniotic fluid stem cells), and angiogenic properties (as measured using chicken embryo assays).
A series of papers have described the development and use (again mainly for tissue engineering purposes) of collagen-based scaffolds that resemble the 3D structure of the finger-like intestinal villi (75,76,82,85), such as the ones present in ileum and jejune. For example, Costello et al. developed an in vitro artificial intestine from poly(lactic-co-glycolic acid) (PLGA) (69,70,86), which could be molded into villous shapes to mimic the topography of the small intestine, thereby providing a platform for the differentiation of epithelial cell types and the subsequent adhesion/invasion of pathogenic bacteria (Figure 2D). The authors showed that different strains of bacteria could attach to epithelial cells residing at different locations of the crypt-villus axis. In addition, in this 3D environment, probiotics exerted their effects through different mechanisms. For example, Lactobacillus gasseri was more effective at displacing Salmonella typhimurium once it had colonized, whereas E. coli Nissle was more effective at preventing attachment of S. typhimurium (Figure 2E). Alternatively, direct 3D-bioprinting has been used to fabricate crypt-villi structures with some degree of tissue functionality. Kim et al. (76) developed a Caco-2 cell-laden collagen ink (cross-linkable with tannic acid), and 3D-bioprinted villi-like structures in which expression of three indicators of epithelial differentiation [MUC17, E-cadherin, and alkaline phosphatase (ALP)] was observed (Figure 2F).
Another important consideration regarding the development of gut-on-chip systems is the type of mammalian cells to use to mimic the host cell system. Most existing in vitro models of human intestinal function commonly rely on the use of established epithelial cell lines, such as Caco-2 (87,88) and HT29-MTX (89) cells. Caco-2 cells are perhaps the best-established cell model for fabricating polarized epithelial monolayers capable of emulating the architecture and function of the small intestine epithelial membrane, despite their colonic origin (87,90). They have been widely used to conduct absorption studies and drug testing in Transwell systems. Recently, Kim et al. showed that a microfluidic gut-on-chip technology that exposed cultured cells to physiological peristalsis-like motions and liquid flow could be used to induce human Caco-2 cells to undergo a spontaneous and robust morphogenesis to produce 3D intestinal villi-like structures (68,91). The cells that formed these villus structures were linked by tight junctions and covered by brush borders (micro-villi-like structures) and produced mucus. They also reconstituted basal proliferative crypts that populated the villi along the crypt-villus axis and formed four different types of differentiated epithelial cells (absorptive, mucus-secretory, enteroendocrine, and Paneth cells) that took characteristic positions similar to those observed in the human small intestine.
Formation of these intestinal villi also resulted in increased intestinal surface area that mimicked the absorptive efficiency of the human intestine, as well as enhanced cytochrome P450 3A4 isoform-based drug metabolizing activity, when compared to conventional Caco-2 cell monolayers cultured in a static transwell system. The ability of the human gut-on-chip to recapitulate the 3D structures, differentiated cell types, and multiple physiological functions of normal human intestinal villi indicate that it may become a powerful alternative in vitro model for studies on intestinal physiology and digestive diseases, as well as an appropriate drug development/screening platform. Cellular models, based on the use of Caco-2, mucus-producing HT29 cells, and Raji B cells have been proposed for a better recapitulation of the dynamics of drug absorption in the small intestine (92,93). Alternatively, HeLa cells, a cervical cancer cell line, have been also used as model cells in simple gut-on-chip systems (67). In a recent contribution, Kasendra et al. developed a small intestine-on-chip device using biopsy-derived organoids (94). The ability of human pluripotent cells to develop fully functional and self-renewable intestinal tissue was also recently demonstrated by Spence et al. (95).
Microbiota-related studies in gut-on-chip systems
A wide variety of in vitro systems have been used to recreate some of the host-microbiota interactions occurring in the human intestine (32,39,69,96-98) or interactions between microbiota protagonists (99). In this section, we have selected and will review some of the systems that we consider closest to the concept of a gut-microbiota-on-chip: a gut-on-chip system in which the interplay of (at least) one bacterial component of human intestinal microbiota and (at least) one human host cell is studied (67,68,71,72,100). We discuss different devices designed to recreate relevant features of the interactions between the host and the human microbiome. We follow a rationale of increasing complexity; we first describe simple systems in which biological or chemical environments appropriate to the human intestine are recreated and one human cell type is exposed to them. Subsequently, we discuss devices in which the physical environment of the gut is the scenario for the interaction of one or more host cells, and one or more microbiota microorganisms. Then, we describe more complex platforms in which human cells and more than two protagonists of the human microbiota interact.
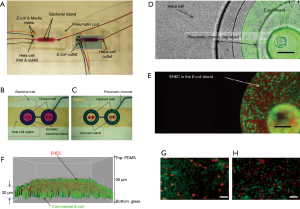
Kim et al. described the co-culture of epithelial cells and bacteria for investigating host-pathogen interactions (67). In particular, the authors aimed to reproduce the sequence of events in gastrointestinal infection in which planktonic pathogen cells first interact with a commensal bacterial biofilm to later attach to epithelial cells. They used a pneumatically actuated system (Figure 3A) composed of two chambers for independent cultures of HeLa cells (cervical cancer cells taken as a model of epithelial intestinal cells) and commensal bacteria (an E. coli BW25113 strain). The HeLa cells were cultured to confluence, while the bacteria were cultured in confined areas (bacterial islands) as a bacterial biofilm (Figure 3B). Each bacterial island (1,200 µm in diameter and 1,000 µm apart) had a separate inlet and outlet for providing growth media and removing waste from the island (Figure 3C). At a later time, the authors emulated a process of infection of the intestinal epithelial cells by adding the enterohemorrhagic and pathogenic E. coli O157:H7 (CDC EDL933 or EHEC) to the co-culture model. For this purpose, the authors first seeded the pathogenic E. coli strain into the commensal bacterial biofilm region and incubated for several hours (Figure 3D). These invasive bacteria were not allowed to install at the commensal bacterial biofilm, and instead chemically interacted with it as planktonic cells (Figure 3E,F). Communication between the HeLa cell zones and the bacterial islands was then permitted, and the viability of the HeLa cells was monitored (Figure 3G,H). The findings suggested that the interaction between the chemical biofilm of the commensal bacteria and the invasive bacteria was key to determining the course of infection. The degree of infection, as measured by the viability of HeLa cells exposed to EHEC, was lower when the invading bacteria interacted with naive commensal bacteria (indole producers) than when they interacted with mutant E. coli lacking indole expression. This paper elegantly shows that the commensal biofilm microenvironment may be a key determinant controlling infectivity and virulence.
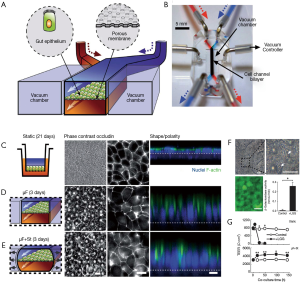
A series of articles by Kim and Ingber have described the evolution of a gut-on-chip system to study different aspects of the host-microbiota interaction in the human intestine. In a first report, Kim and Ingber developed a biomimetic ‘human gut-on-chip’ microdevice aimed at studying the effect of peristalsis and shear on the intestinal epithelial cell layer (72). The architecture of this system more closely resembled that of the human intestine (Figure 2A). The authors also aimed to reproduce the dynamic flow environment of human intestine within this gut-on-chip model. The system was composed of two microfluidic channels separated by a porous flexible membrane coated with extracellular matrix (containing 50 mg/mL of rat type I collagen and 300 mg/mL of Matrigel® in serum-free Dulbecco Modified Eagle Medium (DMEM) and lined with human intestinal epithelial cells (Caco-2, Figure 4A). The gut-on-chip system was composed of clear polydimethylsiloxane (PDMS) elastomer, and a syringe pump was used to perfuse culture media (direction indicated by arrows) through tubing to the upper and lower microchannels (Figure 4B). The gut microenvironment was recreated by flowing culture medium at a low flow rate (0.5 µL/min) and producing low shear stress (0.02 dynes/cm2). In addition, the authors imposed cyclic strain (10%; 0.15 Hz) that mimicked the physiological peristaltic motion. The Caco-2 cells under peristaltic motion underwent phenotypic and expression modifications (Figure 4C,D,E) and assumed some of the key features of the structure of the intestinal epithelium, including the development of villi and crypt-like structures, tight junctions sealed with occludin, cell polarization, and increased height of the cell layer when compared to cells in transwell culture systems (Figure 4C,D,E). The authors also assessed the ability of their system to sustain bacterial growth. Lactobacillus rhamnosus GG (LGG), a habitual intestinal microbe, was co-cultured on the luminal surface of the cultured epithelium for seven days with no compromising of the epithelial cell viability. Growth of these probiotic bacteria improved the barrier function, as observed in the human gut (Figure 4F,G). This gut-on-a-chip recapitulated multiple dynamic physical and functional features of the human intestine. For the first time, shear flow on the intestinal epithelial cell layer and peristaltic motion were integrated into the design of a gut-on-chip system (72). These two factors are critical for the proper recapitulation of intestinal transport phenomena and absorption within a controlled microfluidic environment.
In a more recent contribution, Kim et al. reported a human gut-on-chip platform and its use to study interactions between components of the intestine, the immune system, and bacteria (68). This system was not only able to mimic intestine function (mechanical peristalsis, luminal flow) and tissue architecture, but it was also capable of recreating, for the first time, inflammatory events, with results that recreated in vivo situations and behaviors. The authors used their microbiota-on-chip system to reproduce different scenarios that involved several relevant actors of inflammatory bowel disease, namely immune cells, commensal bacteria, pathogenic bacteria, probiotic bacteria, bacterial endotoxin, cytokines, and antibiotics. They challenged the gut tissue with different stimuli (individual and combined) and assessed their effects on villus integrity, epithelial barrier function, and inflammatory responses. Their results showed the occurrence of villus injury and a detrimental effect in the intestinal barrier integrity when the system was fed with immune cells, i.e., peripheral blood mononuclear cells (PBMCs). A similar effect was observed in the presence of (commensal and/or pathogenic) bacteria and PBMCs. The villus architecture and the intestinal barrier were also compromised in the presence of a bacterial endotoxin, namely lipopolysaccharide (LPS) and immune cells (Figure 5A).
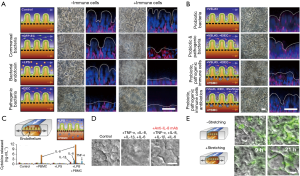
The authors also replicated the well-known effect observed with probiotic bacteria and antibiotics against pathogenic bacteria. Their results indicated that the intestinal villus injury caused by pathogenic bacteria in presence of immune cells could be prevented by the administration of probiotics and antibiotics (Figure 5B). Interestingly, this platform was capable of recapitulating the cytokine secretion profile of vascular endothelial cells when challenged with inflammatory agents, such as LPS, in the presence of immune cells (Figure 5C). In this scenario (LPS + immune cells), four pro-inflammatory cytokines, interleukin (IL)-1β, IL-6, IL-8, and tumor necrosis factor (TNF-α), were secreted at significantly higher concentrations than observed in the control (without immune cells nor LPS).
In a different set of experiments, the authors evaluated the ability of these four proinflammatory cytokines to cause villi injury per se (Figure 5D). They observed that villus damage only occurred when the four cytokines were combined and administered to the system; the cytokines tested individually did not exert this effect. Figure 5D also shows that the intestinal villus damage was prevented when a monoclonal antibody against IL-8 was co-administered with the cocktail of cytokines. Bacterial overgrowth is an important scenario to reproduce and study because it is a characteristic often observed in different intestinal pathologies. In the same work, Kim et al. found that halting the peristalsis-like movement in the gut-on-chip triggered rapid bacterial overgrowth. In a 21-day experiment, the bacterial density increased more than two-fold in the system with no peristaltic stimulation when compared to a system with mechanical stretching (Figure 5E). The authors then studied living human intestinal epithelium, with or without vascular and lymphatic endothelium, immune cells, and mechanical deformation, as well as the living microbiome and pathogenic microbes, and they identified previously unknown contributions of specific cytokines, mechanical motions, and the microbiome to intestinal inflammation, bacterial overgrowth, and control of the barrier function. This contribution by Kim et al. (68) describes in detail the development of a micro-engineered model of human intestinal inflammation and bacterial overgrowth that enables the analysis of individual contributors to the pathophysiology of intestinal diseases, such as ileus and inflammatory bowel disease, over a period of several weeks.
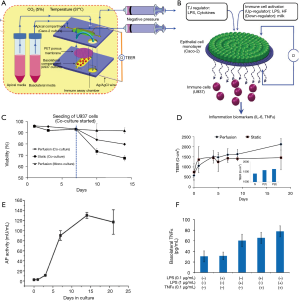
Another relevant angle of application of microbiota gut-on-chip systems is the understanding of the relationships of human nutrition and human microbiota. Ramadan et al. introduced the concept of the NutriChip, an integrated microfluidic platform for investigating the response of immune cells to pro-inflammatory stimuli or immunomodulators in food (37,101). The NutriChip was a miniaturized artificial human gastrointestinal tract, which consisted of two main culture chambers (Figure 6A). The upper chamber hosted a confluent monolayer of epithelial cells that interacted through a permeable membrane with immune cells (monocytes differentiated into macrophages) cultured in the lower compartment (Figure 6B,C). The authors characterized the integrity of the monolayer of Caco-2 cells by trans-epithelial electrical resistance (TEER) measurements (Figure 6D) and conducted across-membrane transport studies using 4-kDa fluorescein isothiocyanate (FITC)-dextran. In addition, the proper differentiation of the cells into intestinal epithelial-like cells was validated by measuring ALP activity (i.e., the cells’ ability to convert p-nitrophenylphosphate into phosphate and p-nitrophenol, Figure 6E). The cell monolayer was confluent after 1 week of microfluidic culture, and the cells were fully differentiated after 2 weeks of culture. In two separate reports (37,101), macrophages (differentiated from U937 monocytes) were exposed to different concentrations of LPS or TNF-α and the associated production of a pro-inflammatory signal, namely IL-6, was measured using a magnetic bead-based immunoassay (Figure 6F). Through this strategy, the authors provided proof-of-concept evidence for the ability to study pro-inflammatory effects of food in a gut-on-chip system. The aim of the NutriChip was to enable the study of processes inherent in the passage of nutrients though the human gut, including, in particular, the activation of immune cells in response to the transfer of nutrients across the epithelial layer. The NutriChip platform offered a new option to evaluate the influence of food quality on health, by monitoring the expression of relevant immune cell biomarkers.
Marzorati et al. developed a microfluidic system—the host-microbiota interaction (HMI) module—to study the response of a monolayer of Caco-2 cells challenged with metabolic products derived from the activity of bacterial biofilms (56). The HMI module was composed of two flow chambers divided by a semipermeable membrane (Figure 7A). The top chamber was fed with bacteria [either a suspension of L. rhamnosus or a complex mix of microorganisms developed and fed from a Simulator of the Human Intestinal Microbial Ecosystem (SHIME)] (Figure 7B). A semipermeable membrane, coated with porcine mucin, divided both chambers and served both as a scaffold to support the development of a bacterial biofilm and as a semi-permeable barrier to allow the diffusion of soluble materials from the bacterial chamber to the bottom chamber. The bottom chamber was continuously fed with mammalian cell culture to maintain a confluent monolayer of Caco-2 cells attached to a fibronectin-coated glass slide.
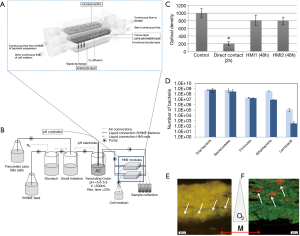
In a first set of experiments, a suspension of L. rhamnosus was continuously fed to the upper chamber. A bacterial biofilm was established in the mucin layer after 1.5 hours of circulation of the bacterial suspension. Similarly, in a second set of experiments, the upper chamber was fed with a complex mix of bacteria developed in the SHIME simulator to establish a bacterial biofilm community. The bacterial and mammalian cell culture media were then continuously fed to the upper and lower chamber, respectively, for 48 hours so that the Caco-2 cell monolayer was indirectly exposed to the products of the bacterial biofilm. The results indicated that the viability of Caco-2 cells at the monolayer was not compromised during this time period. By contrast, Caco-2 cells directly exposed to the complex bacterial mix (in an independent well-plate experiment) exhibited high mortality within the same time frame (Figure 7C).
The authors further studied the composition of the bacterial biofilm after 24 and 48 hours of culture (Figure 7D) and demonstrated that particular strains of bacteria would migrate and establish in different locations (at different heights) of the mucin layer, according to their particular oxygen needs. For instance, Faecalibacterium prausnitzii, a micro-aerophilic bacterium, settled at the basal section of the semipermeable membrane, while Bifidobacterium spp., a strict anaerobe, established in the lumen side of the mucin layer, which was the zone with the lowest oxygen concentration (Figure 7E,F).
In a third set of experiments, Marzorati et al. demonstrated that the mucus-associated microbiota composition was influenced by treatment with a dried product derived from yeast fermentation, a formulation with alleged anti-inflammatory properties. The decreased levels of IL-8 produced by the Caco-2 cell monolayer in these experiments suggested that the HMI was indeed able to recreate the in vivo anti-inflammatory effect attributed to the yeast product. The HMI module aimed to recapitulate several conditions that were physiologically relevant for the gastrointestinal tract, including the presence of a mucosal area, the action of physiological shear stress values (3 dynes/cm2); the bilateral transport of low molecular weight metabolites (4 to 150 kDa) with permeation coefficients ranging from 2.4×10−6 to 7.1×10−9 cm/sec; and the microaerophilic conditions at the bottom of the growing biofilm. The HMI module offered the possibility of co-culturing a gut representative microbial community with enterocyte-like cells for up to 48 hours. To our knowledge, this contribution by the Marzorati Group is the most illustrative example of the coculture and chemical interaction of intestinal cells with a complex mix of intestinal bacteria in a gut-on-chip system.
More recently, Shah et al. developed the human-microbial crosstalk (HuMiX) model (71), a well-designed microbiota-gut-on-chip system that enables co-culture of human and microbial cells under conditions representative of the gastrointestinal human–microbe interface (Figure 8A). The device was composed of a modular stacked assembly of three elastomeric gaskets (thickness: 700 mm) sandwiched between two polycarbonate (PC) enclosures. Each gasket defined a distinct spiral-shaped microchannel 200-mm long, 4-mm wide, and 0.5-mm high. Semi-permeable membranes affixed to the elastomeric gaskets demarcated three channels of 400 mL each. The top microchannel (microbial microchamber) contained a bacterial biofilm. The middle microchannel (human chamber) hosted Caco-2 cells growing on a membrane in a monolayer fashion, while the bottom channel (perfusion chamber) functioned to perfuse culture media to the human microchamber through a porous membrane (Figure 8B). Each microchamber had dedicated inlets and outlets to inject laminar steams of distinct liquids, thereby providing the means for tight control of a distinct cell microenvironment in each chamber and for collecting samples for downstream analysis. The pore sizes of the membranes were chosen for their intended functionality. A microporous membrane (pore diameter of 1 mm), which allows diffusion-dominant perfusion to the basal side of the Caco-2 cells, was used to partition the perfusion and human microchambers; this configuration emulates the intestinal blood supply to the epithelial intestine layer and provides physiologically relevant shear-free culture conditions. A nanoporous membrane (pore diameter of 50 nm) partitioned the human and microbial microchambers to prevent the infiltration of microorganisms, including viruses, into the human microchamber, thereby mimicking the functionality of an intact epithelial barrier.
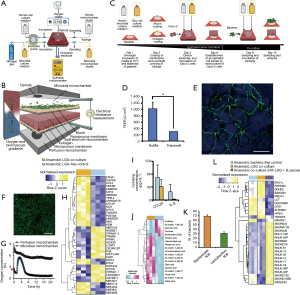
The authors described a step-wise protocol for the coculture of bacterial and human cells in the HuMIX, with negligible loss of viability of the Caco-2 cells (Figure 8C). First, Caco-2 cells were seeded and cultured for 1 week on the mucin and collagen-coated porous membrane to achieve a confluent monolayer culture. The integrity of the epithelial cell barrier was characterized by TEER evaluations (Figure 8D) and observation of the presence of occludin cell junctions by immunofluorescence assays (Figure 8E). A culture of the facultative anaerobe L. rhamnosus (LGG) was then established in the microbial chamber (Figure 8F). LGG, a well-known and characterized lactic acid-producing probiotic, was selected by the authors as a representative member of the Firmicutes phylum. A stream of nitrogen was used to adjust the initial oxygen concentration level of the culture media supplied to the microbial chamber to facilitate the establishment of aerobic or anaerobic environments within it. The system also had optodes (oxygen electrodes) embedded for continuous monitoring of oxygen concentrations in the bacterial chamber (Figure 8G).
The authors demonstrated the ability of the HuMIX to recapitulate the in vivo transcriptional, metabolic, and immunological responses in human intestinal epithelial cells following their co-culture with the commensal LGG grown under anaerobic conditions (Figure 8H,I,J). For instance, they found more than 200 genes that were differentially over- or under-expressed in the HuMIX LGG-Caco-2 cocultures versus HuMIX LGG-free Caco-2 cell cultures (Figure 8H). Moreover, the transcriptomic results obtained in HuMIX cocultures exhibited a high level of agreement with a previously reported analysis of in vivo transcriptomic data from experiments in human volunteers administered LGG. Remarkably, the authors also found that the set of differentially expressed genes in the LGG-Caco-2 cultures was significantly different if aerobic or anaerobic conditions were established in the microbial chamber.
In addition, the authors demonstrated the stable co-culture of LGG and with the obligate anaerobe Bacteroides caccae in the microbial chamber under strict anaerobic conditions. After several days of culture, the bacteria in the microbial chamber reached equilibria in terms of both dissolved oxygen concentration and population composition (~30% B. caccae and ~70% L. rhamnosus, Figure 8K). Co-culture of Caco-2 cells with B. caccae and LGG resulted in a transcriptional response that was distinct from that of a co-culture solely comprising LGG (Figure 8L). Systems like the HuMiX will facilitate fundamental research of host–microbe molecular interactions and will provide insights into a range of research questions linking the composition of gastrointestinal microbiota to human health and disease.
This first set of contributions on microbiota-on-chip systems has mainly consisted of proof-of-concept studies. Nevertheless, these papers present clear and solid evidence supporting the great potential of gut microbiota-on-chip models for studying and understanding complex physiological processes (i.e., the role of peristalsis on the intestinal dynamics or the competition between commensal and pathogenic bacteria). Further expansion of the use of microbiota-on-chips to other applications (i.e., modeling of disease, evaluation of the effect of tensors on microbiota balance, nutrition, and probiotic-on-chip applications) will require that some important challenges first be overcome. In our view, these challenges are mainly related to limitations on the portfolio of microfabrication techniques and biomaterials currently available, the need for more complex bacterial-cell interaction models, and the need for sensing and instrumentation platforms that are fully adapted to organ-on-chip applications. Next, we briefly discuss specific challenges, opportunities, and main enablers of the next generation of gut microbiota-on-chip platforms.
Challenges, opportunities, and enabling technologies for the next generation of gut-microbiota-on-chip devices
The next generation of gut microbiota-on-chip systems will be enabled by technologies such as 3D printing/bioprinting, microfluidics, and microfabrication, as well as by stem cell engineering. These innovative technologies are anticipated to speed up and simplify the development of better microbiota-on-chip models.
Today, PDMS and poly(methyl methacrylate) (PMMA), continue to be the most widely used materials for building lab-on-chips. The design of most organ-on-chip devices typically requires the assembly of multiple layers of these two materials, which implies the need for laborious work to eliminate hydraulic leaks. To this end, 3D printing is an attractive alternative to the use of multi-layered PDMS/PMMA devices. The cost-effective and convenient use of 3D printing to fabricate microfluidic devices has been widely documented in recent literature (102,103), including a report relative to the fabrication of a gut-on-chip microreactor (86). The ongoing parallel evolution of microfluidics and novel materials with attractive characteristics (transparency, temperature resistance, reusability) will further expand the portfolio of materials available for 3D printing (104). For example, the use of high-resolution stereolithographic 3D printing techniques can generate highly complex microfluidic devices at a small fraction of the cost per device and greatly reduced fabrication time when compared with silicon-based counterparts (105). These printing techniques will greatly expedite and simplify the process of fabricating gut-on-chip (and other organ-on-chip) systems.
Recent evidence suggests that not only the diverse composition, but also the spatial distribution, of bacterial communities in the intestine determines the functionality of the human gut microbiome (94). New technologies for bioprinting bacteria and bacterial communities (49,106,107), will facilitate the predesigned deposition of specific strains, allowing the development of specific architectures and the control of the degree of interaction of “engineered” microbiota communities. Bioprinting will also facilitate the fabrication of cell-laden constructs that resemble the architecture of the intestinal membrane (76). In this regard, novel multi-material bioprinting techniques (108) offer the possibility of bioprinting different bacterial strains and human cells in close proximity and in predetermined patterns.
The intestinal epithelium is the most rapidly self-renewing tissue in adult mammals. New stem cell technologies may also provide additional tools to develop epithelial intestinal micro-tissues with self-renewal capabilities for microbiota-on-chip applications (109). For example, the use of human induced pluripotent stem cells (iPSCs) to engineered intestinal membranes with self-renewal capabilities appears to be feasible. Spence et al. established an efficient process to direct the differentiation of human iPSCs into intestinal tissue in vitro using a temporal series of growth factor manipulations to mimic embryonic intestinal development (95). Alternatively, Sato et al. recently demonstrated that single sorted Lgr5+ stem cells can form intestinal crypt villus organoids (110). Patil et al. demonstrated that bone marrow stem cells can repopulate decellularized intestine (84). Stem cell technologies will certainly improve the available methodologies for fabricating physiologically meaningful intestinal membranes.
The presence of the mucus layer has been neglected in most gut-on-chip devices reported so far. Caco-2 cells, the most frequently used epithelial cell model for gut-on-chip systems, do not secrete mucus under static conditions but produce mucus (not fully characterized) under flow and peristaltic stimulation (68). The proper mimicry of the mucus layer and its properties will add realism to the emulation of chemical and physical interactions between the bacterial biofilm and the monolayer of epithelial cells in the human intestine. After all, under normal circumstances, bacteria and human cells do not interact directly, but only through the mucus layer. Mucus is composed of ~50 mucins, with Muc2 being the most abundant among them (80,111). Muc2 (NCBD; gene ID: 4583) is a high-molecular weight and highly glycosylated protein. Its structural complexity and size have made recombinant production of Muc2 a challenge (112). Although some mucin and mucin-like proteins are commercially available, the engineering of protein-based hydrogels (79,89) is a clear niche of future research. We foresee that hydrogels or mucin-like formulations will be frequently used in near-future versions of gut-on-chip systems. Hydrogel-based materials (113,114) with enhanced functionalities (i.e., spatial-temporal control of characteristics) can be tailored to match some of the relevant properties and functions of intestinal mucin for specific applications (115); these novel materials will enrich the arsenal of options available to fine-tune the properties of the mucus layer in gut-on-chips.
Final remarks
Understanding the complex functioning of gut microbiota and its ad hoc modification has tremendous potential to improve health and prevent some of the diseases that are of the most concern today in public health. For instance, diabetes, hypertension, and cancer are probably the three most concerning “epidemics” of modern society. These three complex health problems have been, at least in part, associated with microbiota dysbiosis (7,10,12,25,27). Microbiota-gut-on-chip systems will be key players in developing and testing therapies for all these life-threatening disorders.
The continuous culture of parasites that are difficult to expand by conventional culture techniques is another field in which gut-on-chips have tremendous potential (116). For example, the development of effective drugs against Cryptosporidium infection has been hindered by the lack of culture systems capable of recapitulating most of the stages of the complex life cycle of the parasite. The expansion of Cryptosporidium sp. (and other parasites) requires the presence of mammalian cells to recreate the processes of cellular infection and the occurrence of the intracellular stages of its reproductive cycle (117). Although some in vitro models of intestine organoids have been used to study Cryptosporidium infection (117,118), no gut-on-chip model has yet been used for the continuous culture of this host-dependent parasite.
The recreation and extended control of anaerobic and/or microaerophilic conditions is another challenge in the engineering of microbiota gut-on-chip systems (38,71,117,119) for these applications. In general, by perfecting the control of the microenvironment in a microfluidic system, gut-microbiota-on-chips will become a valuable tool for a more comprehensive study and treatment of long-term affections such as parasite infection, Helicobacter pylori chronic infection (120), chronic ulcerative processes, or celiac disease (121-123). The culture of “unculturable” (recalcitrant) bacteria (48,51) is another obvious topic of application of gut-on-chips. These systems have a full potential to enable the proper recreation of the different chemical, physical, and ecological environments that prevail in different sections of the human intestine and will certainly change the definition of “difficult to culture” microbial species.
In addition, the development of better gut-microbiota-on-chip models capable of reproducing the mechanical, structural, absorptive, transport, and pathophysiological properties of the human gut, in synchrony with its crucial microbial symbionts, could accelerate pharmaceutical development, and potentially replace or certainly minimize and optimize animal testing (72,124). Microbiota-on-chip assays will also make important contributions to our understanding of the effects of the food we consume (37,56), the balance of our microbiota, and our overall health.
However, microbiota-on-chip technologies face important challenges to fulfill all these high goals. An effective use of gut-on-chip systems to evaluate the absorption rate for pharma compounds and nutrients from food will demand a more precise reproduction of important parameters of the intestine, such as effective permeability (124,125) to specific compounds and surface area (124,126). The precise estimation of these parameters already presents experimental challenges. An additional complication is that the values of these parameters differ significantly among different individuals and throughout different regions of the intestinal tract (124-126). On top of this, a more rigorous identification of key factors influencing the delicate balance in microbiota composition is required. For example, a recent study showed that different variables, such as local pH values, water flow, and the intestinal microenvironment determine the composition of human microbiota (46). Flow conditions in the intestine are particular and dynamic (77,91,127), and intestinal contractive activity, including the occurrence of peristaltic waves, plays an important role in defining these flow conditions and contributing to enhanced mixing, material transport, and even host-cell morphology and differentiation (68,91). Indeed, mixing in the intestinal system is a multi-scale process. At the level of the mucosa, small villi effectively contribute to mixing of materials in a laminar layer (77) and enhance mass transfer of nutrients, enzymes, and other active players in the absorption process in the neighborhood of the endothelial cell layer. Computational fluid dynamic (CFD) simulations have been used to understand and model flow condition in the human intestine and in a few gut-on-chip systems (86,101). In the years to come CFD will certainly become a key player on the design on microbiota-on-chip systems.
The sophistication and robustness of the next generation of gut-on-chips should enable a much better understanding of the interplay between the human microbiota and human physiology. In the years to come, gut-on-chip systems will play a key role in solving important questions related to the overall health in humans, including the nutritional effects of food, the influences of food additives and specific pharma compounds, and the etiology, diagnostics, and treatment of specific diseases, among others. New microfabrication techniques, novel biomaterials, improved sensing and instrumentation strategies [i.e., mini-microscopes (128,129) and on-line sensors (130)], and more complex bacterial-cell systems should be incorporated to accomplish these high aims.
Acknowledgments
Funding: G Trujillo-de Santiago and MM Alvarez gratefully acknowledge the financial support of Consejo Nacional de Ciencia y Tecnología (CONACYT), and the Strategic Research Iniciative of Tecnológico de Monterrey, México. YS Zhang acknowledges the funding from the National Institutes of Health (K99CA201603, R21EB025270) and the New England Anti-Vivisection Society (NEAVS).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/mps.2018.09.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hooper L V, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol 2010;10:159-69. [Crossref] [PubMed]
- Clemente JC, Ursell LK, Parfrey LW, et al. The Impact of the Gut Microbiota on Human Health: An Integrative View. Cell 2012;148:1258-70. [Crossref] [PubMed]
- Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol 2014;12:661-72. [Crossref] [PubMed]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207-14. [Crossref] [PubMed]
- Lynch SV, Pedersen O. The Human Intestinal Microbiome in Health and Disease. N Engl J Med 2016;375:2369-79. [Crossref] [PubMed]
- Karkman A, Lehtimäki J, Ruokolainen L. The ecology of human microbiota: dynamics and diversity in health and disease. Ann N Y Acad Sci 2017;1399:78-92. [Crossref] [PubMed]
- Garrett WS. Cancer and the microbiota. Science 2015;348:80-6. [Crossref] [PubMed]
- Logan AC, Jacka FN, Prescott SL. Immune-Microbiota Interactions: Dysbiosis as a Global Health Issue. Curr Allergy Asthma Rep 2016;16:13. [Crossref] [PubMed]
- Joossens M, Huys G, Cnockaert M, et al. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut 2011;60:631-7. [Crossref] [PubMed]
- Tilg H, Moschen AR. Microbiota and diabetes: an evolving relationship. Gut 2014;63:1513-21. [Crossref] [PubMed]
- Larsen N, Vogensen FK, Van Den Berg FWJ, et al. Gut Microbiota in Human Adults with Type 2 Diabetes Differs from Non-Diabetic Adults. PLoS One 2010;5:e9085 [Crossref] [PubMed]
- Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012;490:55-60. [Crossref] [PubMed]
- Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nat Immunol 2011;12:5-9. [Crossref] [PubMed]
- Lee YK, Menezes JS, Umesaki Y, et al. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A 2011;108:4615-22. [Crossref] [PubMed]
- Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 2009;9:313-23. [Crossref] [PubMed]
- Trompette A, Gollwitzer ES, Yadava K, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med 2014;20:159-66. [Crossref] [PubMed]
- Dzidic M, Abrahamsson TR, Artacho A, et al. Aberrant IgA responses to the gut microbiota during infancy precede asthma and allergy development. J Allergy Clin Immunol 2017;139:1017-25.e14. [Crossref] [PubMed]
- Frank DN, St Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 2007;104:13780-5. [Crossref] [PubMed]
- Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature 2007;448:427-34. [Crossref] [PubMed]
- Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature 2009;457:480-4. [Crossref] [PubMed]
- Gao R, Zhu C, Li H, et al. Dysbiosis Signatures of Gut Microbiota Along the Sequence from Healthy, Young Patients to Those with Overweight and Obesity. Obesity 2018;26:351-61. [Crossref] [PubMed]
- Kim KA, Gu W, Lee IA, et al. High Fat Diet-Induced Gut Microbiota Exacerbates Inflammation and Obesity in Mice via the TLR4 Signaling Pathway. PLoS One 2012;7:e47713 [Crossref] [PubMed]
- Vijay-Kumar M, Aitken JD, Carvalho FA, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 2010;328:228-31. [Crossref] [PubMed]
- Chassaing B, Etienne-Mesmin L, Gewirtz AT. Microbiota-liver axis in hepatic disease. Hepatology 2014;59:328-39. [Crossref] [PubMed]
- Vipperla K, O’Keefe SJ. Diet, microbiota, and dysbiosis: a ‘recipe’ for colorectal cancer. Food Funct 2016;7:1731-40. [Crossref] [PubMed]
- Arthur JC, Perez-Chanona E, Mühlbauer M, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012;338:120-3. [Crossref] [PubMed]
- Marques FZ, Mackay CR, Kaye DM. Beyond gut feelings: how the gut microbiota regulates blood pressure. Nat Rev Cardiol 2018;15:20-32. [Crossref] [PubMed]
- Neville BA, Forster SC, Lawley TD. Commensal Koch’s postulates: establishing causation in human microbiota research. Curr Opin Microbiol 2018;42:47-52. [Crossref] [PubMed]
- Turnbaugh PJ, Ley RE, Hamady M, et al. The human microbiome project. Nature 2007;449:804-10. [Crossref] [PubMed]
- Sampson TR, Debelius JW, Thron T, et al. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson’s Disease. Cell 2016;167:1469-80.e12. [Crossref] [PubMed]
- Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010;464:59-65. [Crossref] [PubMed]
- Arnold JW, Roach J, Azcarate-Peril MA. Emerging Technologies for Gut Microbiome Research. Trends Microbiol 2016;24:887-901. [Crossref] [PubMed]
- Mark Welch JL, Hasegawa Y, McNulty NP, et al. Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. Proc Natl Acad Sci U S A 2017;114:E9105-14. [Crossref] [PubMed]
- Hugenholtz F, de Vos WM. Mouse models for human intestinal microbiota research: a critical evaluation. Cell Mol Life Sci 2018;75:149-60. [Crossref] [PubMed]
- Nguyen TLA, Vieira-Silva S, Liston A, et al. How informative is the mouse for human gut microbiota research? Dis Model Mech 2015;8:1-16. [Crossref] [PubMed]
- Dosh RH, Jordan-Mahy N, Sammon C, et al. Tissue Engineering Laboratory Models of the Small Intestine. Tissue Eng Part B Rev 2018;24:98-111. [Crossref] [PubMed]
- Ramadan Q, Jafarpoorchekab H, Huang C, et al. NutriChip: nutrition analysis meets microfluidics. Lab Chip 2013;13:196-203. [Crossref] [PubMed]
- Shaban L, Chen Y, Fasciano AC, et al. A 3D intestinal tissue model supports Clostridioides difficile germination, colonization, toxin production and epithelial damage. Anaerobe 2018;50:85-92. [Crossref] [PubMed]
- Park GS, Park MH, Shin W, et al. Emulating Host-Microbiome Ecosystem of Human Gastrointestinal Tract in Vitro. Stem Cell Rev 2017;13:321-34. [Crossref] [PubMed]
- Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol 2016;14:e1002533 [Crossref] [PubMed]
- Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 2011;108:4578-85. [Crossref] [PubMed]
- Paliy O, Kenche H, Abernathy F, et al. High-throughput quantitative analysis of the human intestinal microbiota with a phylogenetic microarray. Appl Environ Microbiol 2009;75:3572-9. [Crossref] [PubMed]
- Luckey TD. Introduction to intestinal microecology. Am J Clin Nutr 1972;25:1292-4. [Crossref] [PubMed]
- Sender R, Fuchs S, Milo R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell 2016;164:337-40. [Crossref] [PubMed]
- Lozupone CA, Stombaugh JI, Gordon JI, et al. Diversity, stability and resilience of the human gut microbiota. Nature 2012;489:220-30. [Crossref] [PubMed]
- Cremer J, Arnoldini M, Hwa T. Effect of water flow and chemical environment on microbiota growth and composition in the human colon. Proc Natl Acad Sci U S A 2017;114:6438-43. [Crossref] [PubMed]
- Gill SR, Pop M, Deboy RT, et al. Metagenomic analysis of the human distal gut microbiome. Science 2006;312:1355-9. [Crossref] [PubMed]
- Rajilić-Stojanović M, de Vos WM. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev 2014;38:996-1047. [Crossref] [PubMed]
- Lehner BAE, Schmieden DT, Meyer AS. A Straightforward Approach for 3D Bacterial Printing. ACS Synth Biol 2017;6:1124-30. [Crossref] [PubMed]
- Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature 2011;473:174-80. [Crossref] [PubMed]
- Browne HP, Forster SC, Anonye BO, et al. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature 2016;533:543-6. [Crossref] [PubMed]
- Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science 2005;308:1635-8. [Crossref] [PubMed]
- Xu J, Mahowald MA, Ley RE, et al. Evolution of Symbiotic Bacteria in the Distal Human Intestine. PLoS Biol 2007;5:e156 [Crossref] [PubMed]
- Moeller AH, Caro-Quintero A, Mjungu D, et al. Cospeciation of gut microbiota with hominids. Science 2016;353:380-2. [Crossref] [PubMed]
- Rothschild D, Weissbrod O, Barkan E, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018;555:210-5. [Crossref] [PubMed]
- Marzorati M, Vanhoecke B, De Ryck T, et al. The HMITM module: a new tool to study the Host-Microbiota Interaction in the human gastrointestinal tract in vitro. BMC Microbiol 2014;14:133. [Crossref] [PubMed]
- Chassaing B, Koren O, Goodrich JK, et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 2015;519:92-6. [Crossref] [PubMed]
- Pérez-Cobas AE, Gosalbes MJ, Friedrichs A, et al. Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut 2013;62:1591-601. [Crossref] [PubMed]
- Dethlefsen L, Huse S, Sogin ML, et al. The Pervasive Effects of an Antibiotic on the Human Gut Microbiota, as Revealed by Deep 16S rRNA Sequencing. PLoS Biol 2008;6:e280 [Crossref] [PubMed]
- Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature 2012;486:222. [Crossref] [PubMed]
- Mariat D, Firmesse O, Levenez F, et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol 2009;9:123. [Crossref] [PubMed]
- Putignani L, Del Chierico F, Petrucca A, et al. The human gut microbiota: a dynamic interplay with the host from birth to senescence settled during childhood. Pediatr Res 2014;76:2-10. [Crossref] [PubMed]
- Solís G, de los Reyes-Gavilan CG, Fernández N, et al. Establishment and development of lactic acid bacteria and bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe 2010;16:307-10. [Crossref] [PubMed]
- Wiese M, Khakimov B, Nielsen S, et al. CoMiniGut—a small volume in vitro colon model for the screening of gut microbial fermentation processes. PeerJ 2018;6:e4268 [Crossref] [PubMed]
- Payne AN, Zihler A, Chassard C, et al. Advances and perspectives in in vitro human gut fermentation modeling. Trends Biotechnol 2012;30:17-25. [Crossref] [PubMed]
- Rossi E, Cimdins A, Lüthje P, et al. “It’s a gut feeling” – Escherichia coli biofilm formation in the gastrointestinal tract environment. Crit Rev Microbiol 2018;44:1-30. [Crossref] [PubMed]
- Kim J, Hegde M, Jayaraman A. Co-culture of epithelial cells and bacteria for investigating host-pathogen interactions. Lab Chip 2010;10:43-50. [Crossref] [PubMed]
- Kim HJ, Li H, Collins JJ, et al. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci U S A 2016;113:E7-15. [Crossref] [PubMed]
- Costello CM, Sorna RM, Goh YL, et al. 3-D Intestinal Scaffolds for Evaluating the Therapeutic Potential of Probiotics. Mol Pharm 2014;11:2030-9. [Crossref] [PubMed]
- Costello CM, Hongpeng J, Shaffiey S, et al. Synthetic small intestinal scaffolds for improved studies of intestinal differentiation. Biotechnol Bioeng 2014;111:1222-32. [Crossref] [PubMed]
- Shah P, Fritz JV, Glaab E, et al. A microfluidics-based in vitro model of the gastrointestinal human–microbe interface. Nat Commun 2016;7:11535. [Crossref] [PubMed]
- Kim HJ, Huh D, Hamilton G, et al. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 2012;12:2165. [Crossref] [PubMed]
- Esch MB, Sung JH, Yang J, et al. On chip porous polymer membranes for integration of gastrointestinal tract epithelium with microfluidic ‘body-on-a-chip’ devices. Biomed Microdevices 2012;14:895-906. [Crossref] [PubMed]
- Totonelli G, Maghsoudlou P, Garriboli M, et al. A rat decellularized small bowel scaffold that preserves villus-crypt architecture for intestinal regeneration. Biomaterials 2012;33:3401-10. [Crossref] [PubMed]
- Kim SH, Chi M, Yi B, et al. Three-dimensional intestinal villi epithelium enhances protection of human intestinal cells from bacterial infection by inducing mucin expression. Integr Biol (Camb) 2014;6:1122-31. [Crossref] [PubMed]
- Kim W, Kim GH. An innovative cell-printed microscale collagen model for mimicking intestinal villus epithelium. Chem Eng J 2018;334:2308-18. [Crossref]
- Lim YF, de Loubens C, Love RJ, et al. Flow and mixing by small intestine villi. Food Funct 2015;6:1787-95. [Crossref] [PubMed]
- Chen Y, Zhou W, Roh T, et al. In vitro enteroid-derived three-dimensional tissue model of human small intestinal epithelium with innate immune responses. PLoS One 2017;12:e0187880 [Crossref] [PubMed]
- Wang Y, Gunasekara DB, Reed MI, et al. A microengineered collagen scaffold for generating a polarized crypt-villus architecture of human small intestinal epithelium. Biomaterials 2017;128:44-55. [Crossref] [PubMed]
- Birchenough GMH, Johansson ME, Gustafsson JK, et al. New developments in goblet cell mucus secretion and function. Mucosal Immunol 2015;8:712-9. [Crossref] [PubMed]
- Atuma C, Strugala V, Allen A, et al. The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am J Physiol Gastrointest Liver Physiol 2001;280:G922-9. [Crossref] [PubMed]
- Shim KY, Lee D, Han J, et al. Microfluidic gut-on-a-chip with three-dimensional villi structure. Biomed Microdevices 2017;19:37. [Crossref] [PubMed]
- Syed O, Walters NJ, Day RM, et al. Evaluation of decellularization protocols for production of tubular small intestine submucosa scaffolds for use in oesophageal tissue engineering. Acta Biomater 2014;10:5043-54. [Crossref] [PubMed]
- Patil PB, Chougule PB, Kumar VK, et al. Recellularization of Acellular Human Small Intestine Using Bone Marrow Stem Cells. Stem Cells Transl Med 2013;2:307-15. [Crossref] [PubMed]
- Sung JH, Yu J, Luo D, et al. Microscale 3-D hydrogel scaffold for biomimetic gastrointestinal (GI) tract model. Lab Chip 2011;11:389-92. [Crossref] [PubMed]
- Costello CM, Phillipsen MB, Hartmanis LM, et al. Microscale Bioreactors for in situ characterization of GI epithelial cell physiology. Sci Rep 2017;7:12515. [Crossref] [PubMed]
- Hidalgo IJ, Raub TJ, Borchardt RT. Characterization of the human colon carcinoma cell line (Caco-2) as a model system for intestinal epithelial permeability. Gastroenterology 1989;96:736-49. [Crossref] [PubMed]
- Gaillard JL, Berche P, Mounier J, et al. In vitro model of penetration and intracellular growth of Listeria monocytogenes in the human enterocyte-like cell line Caco-2. Infect Immun 1987;55:2822-9. [PubMed]
- Dosh RH, Essa A, Jordan-Mahy N, et al. Use of hydrogel scaffolds to develop an in vitro 3D culture model of human intestinal epithelium. Acta Biomater 2017;62:128-43. [Crossref] [PubMed]
- Yee S. In Vitro Permeability Across Caco-2 Cells (Colonic) Can Predict In Vivo (Small Intestinal) Absorption in Man—Fact or Myth. Pharm Res 1997;14:763-6. [Crossref] [PubMed]
- Kim HJ, Ingber DE. Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr Biol (Camb) 2013;5:1130. [Crossref] [PubMed]
- Antunes F, Andrade F, Araújo F, et al. Establishment of a triple co-culture in vitro cell models to study intestinal absorption of peptide drugs. Eur J Pharm Biopharm 2013;83:427-35. [Crossref] [PubMed]
- Araújo F, Sarmento B. Towards the characterization of an in vitro triple co-culture intestine cell model for permeability studies. Int J Pharm 2013;458:128-34. [Crossref] [PubMed]
- Kasendra M, Tovaglieri A, Sontheimer-Phelps A, et al. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci Rep 2018;8:2871. [Crossref] [PubMed]
- Spence JR, Mayhew CN, Rankin SA, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 2011;470:105-9. [Crossref] [PubMed]
- Marzorati M, Van den Abbeele P, Possemiers S, et al. Studying the host-microbiota interaction in the human gastrointestinal tract: basic concepts and in vitro approaches. Ann Microbiol 2011;61:709-15. [Crossref]
- Everett J, Gabrilska R, Rumbaugh KP, et al. Assessing Pseudomonas aeruginosa Autoinducer Effects on Mammalian Epithelial Cells. Methods Mol Biol 2018;1673:213-25. [Crossref] [PubMed]
- Barrila J, Radtke AL, Crabbé A, et al. Organotypic 3D cell culture models: using the rotating wall vessel to study host–pathogen interactions. Nat Rev Microbiol 2010;8:791-801. [Crossref] [PubMed]
- Luo X, Tsao CY, Wu HC, et al. Distal modulation of bacterial cell–cell signalling in a synthetic ecosystem using partitioned microfluidics. Lab Chip 2015;15:1842-51. [Crossref] [PubMed]
- Huh D, Kim HJ, Fraser JP, et al. Microfabrication of human organs-on-chips. Nat Protoc 2013;8:2135-57. [Crossref] [PubMed]
- Ramadan Q, Jing L. Characterization of tight junction disruption and immune response modulation in a miniaturized Caco-2/U937 coculture-based in vitro model of the human intestinal barrier. Biomed Microdevices 2016;18:11. [Crossref] [PubMed]
- Ho CMB, Ng SH, Li KHH, et al. 3D printed microfluidics for biological applications. Lab Chip 2015;15:3627-37. [Crossref] [PubMed]
- Yazdi AA, Popma A, Wong W, et al. 3D printing: an emerging tool for novel microfluidics and lab-on-a-chip applications. Microfluid Nanofluidics 2016;20:50. [Crossref]
- Hou X, Zhang YS, Trujillo-de Santiago G, et al. Interplay between materials and microfluidics. Nat Rev Mater 2017;2:17016. [Crossref]
- Olvera-Trejo D, Velásquez-García LF. Additively manufactured MEMS multiplexed coaxial electrospray sources for high-throughput, uniform generation of core–shell microparticles. Lab Chip 2016;16:4121-32. [Crossref] [PubMed]
- Connell JL, Ritschdorff ET, Whiteley M, et al. 3D printing of microscopic bacterial communities. Proc Natl Acad Sci U S A 2013;110:18380-5. [Crossref] [PubMed]
- Trujillo-de Santiago G, Alvarez MM, Samandari M, et al. Chaotic printing: using chaos to fabricate densely packed micro- and nanostructures at high resolution and speed. Mater Horiz 2018;5:813-22. [Crossref]
- Liu W, Zhang YS, Heinrich MA, et al. Rapid Continuous Multimaterial Extrusion Bioprinting. Adv Mater 2017;29:1604630 [Crossref] [PubMed]
- Sato T, Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science 2013;340:1190-4. [Crossref] [PubMed]
- Sato T, Vries RG, Snippert HJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009;459:262-5. [Crossref] [PubMed]
- Johansson MEV, Larsson JMH, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A 2011;108:4659-65. [Crossref] [PubMed]
- Bäckström M, Ambort D, Thomsson E, et al. Increased Understanding of the Biochemistry and Biosynthesis of MUC2 and Other Gel-Forming Mucins Through the Recombinant Expression of Their Protein Domains. Mol Biotechnol 2013;54:250-6. [Crossref] [PubMed]
- Yue K, Trujillo-de Santiago G, Alvarez MM, et al. Synthesis, properties, and biomedical applications of gelatin methacryloyl (GelMA) hydrogels. Biomaterials 2015;73:254-71. [Crossref] [PubMed]
- Leijten J, Seo J, Yue K, et al. Spatially and temporally controlled hydrogels for tissue engineering. Mater Sci Eng R Rep 2017;119:1-35. [Crossref] [PubMed]
- Cook MT, Smith SL, Khutoryanskiy VV. Novel glycopolymer hydrogels as mucosa-mimetic materials to reduce animal testing. Chem Commun (Camb) 2015;51:14447-50. [Crossref] [PubMed]
- Aguilar-Rojas A, Olivo-Marin JC, Guillen N. The motility of Entamoeba histolytica: finding ways to understand intestinal amoebiasis. Curr Opin Microbiol 2016;34:24-30. [Crossref] [PubMed]
- Morada M, Lee S, Gunther-Cummins L, et al. Continuous culture of Cryptosporidium parvum using hollow fiber technology. Int J Parasitol 2016;46:21-9. [Crossref] [PubMed]
- Alcantara Warren C, Destura RV, Sevilleja JE, et al. Detection of Epithelial‐Cell Injury, and Quantification of Infection, in the HCT‐8 Organoid Model of Cryptosporidiosis. J Infect Dis 2008;198:143-9. [Crossref] [PubMed]
- von Martels JZH, Sadaghian Sadabad M, Bourgonje AR, et al. The role of gut microbiota in health and disease: In vitro modeling of host-microbe interactions at the aerobe-anaerobe interphase of the human gut. Anaerobe 2017;44:3-12. [Crossref] [PubMed]
- Uemura N, Okamoto S, Yamamoto S, et al. Helicobacter pylori Infection and the Development of Gastric Cancer. N Engl J Med 2001;345:784-9. [Crossref] [PubMed]
- Fasano A, Catassi C. Celiac Disease. N Engl J Med 2012;367:2419-26. [Crossref] [PubMed]
- Rostom A, Murray JA, Kagnoff MF. American Gastroenterological Association (AGA) Institute technical review on the diagnosis and management of celiac disease. Gastroenterology 2006;131:1981-2002. [Crossref] [PubMed]
- Wacklin P, Kaukinen K, Tuovinen E, et al. The Duodenal Microbiota Composition of Adult Celiac Disease Patients Is Associated with the Clinical Manifestation of the Disease. Inflamm Bowel Dis 2013;19:934-41. [Crossref] [PubMed]
- Olivares-Morales A, Lennernäs H, Aarons L, et al. Translating Human Effective Jejunal Intestinal Permeability to Surface-Dependent Intrinsic Permeability: a Pragmatic Method for a More Mechanistic Prediction of Regional Oral Drug Absorption. AAPS J 2015;17:1177-92. [Crossref] [PubMed]
- Lennernäs H. Human in Vivo Regional Intestinal Permeability: Importance for Pharmaceutical Drug Development. Mol Pharm 2014;11:12-23. [Crossref] [PubMed]
- Helander HF, Fändriks L. Surface area of the digestive tract – revisited. Scand J Gastroenterol 2014;49:681-9. [Crossref] [PubMed]
- Lentle RG, De Loubens C, Hulls C, et al. A comparison of the organization of longitudinal and circular contractions during pendular and segmental activity in the duodenum of the rat and guinea pig. Neurogastroenterol Motil 2012;24:686-95, e298.
- Zhang YS, Trujillo-de Santiago G, Alvarez MM, et al. Expansion Mini-Microscopy: An Enabling Alternative in Point-of-Care Diagnostics. Curr Opin Biomed Eng 2017;1:45-53. [Crossref] [PubMed]
- Zhang YS, Chang JB, Alvarez MM, et al. Hybrid Microscopy: Enabling Inexpensive High-Performance Imaging through Combined Physical and Optical Magnifications. Sci Rep 2016;6:22691. [Crossref] [PubMed]
- Zhang YS, Aleman J, Shin SR, et al. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc Natl Acad Sci U S A 2017;114:E2293-302. [Crossref] [PubMed]
Cite this article as: Trujillo-de Santiago G, Lobo-Zegers MJ, Montes-Fonseca SL, Zhang YS, Alvarez MM. Gut-microbiota-on-a-chip: an enabling field for physiological research. Microphysiol Syst 2018;2:7.


