
Microphysiological systems to study microenvironment-cell nucleus interaction: importance of tissue geometry and heterogeneity
Introduction
Three-dimensional (3D) cell culture was developed in the late seventies to demonstrate the importance of the extracellular matrix (ECM) in controlling cell behavior (1-3). After thirty years of nearly absent recognition that 3D cell culture could constitute an unparalleled method to study normal and diseased cell behaviors, the scientific community has placed enormous stress on the field, predicting its invaluable contributions to drug development, precision medicine and organ replacement. This hope is greatly served by new developments in micromanufacturing and the availability of ‘malleable’ cells, such as induced-pluripotent stem cells that finally enable scientists to study tissue complexity and diversity.
Producing microphysiological systems implicitly means obtaining physiologically relevant models of tissues; such is the goal of 3D cell culture overall. From its origins, the definition of this art of cell culture is to place cells in an environment that allows them to organize and behave as they would in vivo. Over the years, it has become clear that, to achieve physiological relevance, it is indispensable to reproduce tissue architecture, namely the essential structural features of a tissue. This requirement was first demonstrated with the mammary epithelium and its basoapical polarity axis in murine and human cell culture models (4-7). Furthermore, the importance of the milieu surrounding cells, or the microenvironment, to mediate tissue architecture was captured when showing that at minimum, laminin 111, a major component of the basement membrane needs to be coated on the cell culture surface for the polarization of luminal epithelial cells in monoculture (6,8). Whereas, collagen I (normally found in the stroma) as the sole ECM leads to inverted polarity of the epithelium (6,7). The existence of ECM receptors at the cell surface, themselves connected to the cytoskeleton that makes contact with the cell nucleus, was the basis for the theory of dynamic reciprocity proposed by Mina Bissell in the early 80s (9), in which nucleus-mediated gene expression and ECM constantly influence each other. As shown over the years, microenvironment-cell nucleus interactions occur at multiple levels. The study of the structural connection between nucleus and ECM is now the booming field of mechanotransduction (10,11) for which the cytoskeleton is a major component. The physical continuity between nucleus and adhesion complexes for cell-cell and cell-ECM connections has been visualized thanks to advanced microscopy techniques (12). Moreover, connection via signal transduction has become increasingly complex, and although still underdeveloped, our proposed idea of connection via the shuttling of resident nuclear proteins to the cytoplasm, in addition to the demonstrated shuttling of resident cytoplasmic proteins to the cell nucleus (13) is gaining momentum (14). Finally, soluble molecules present in the ECM enter cells at different concentrations over time. In addition to the diffusion of molecules from the circulation, local cells release biochemical and genetic information into the ECM via exosomes, indicating that the complexity of microenvironment-nucleus interactions requires well-controlled cell culture systems to further investigate tissue and organ biology.
Our laboratory has participated in the pioneering developments of matrix-related 3D culture with human non-neoplastic and cancer cells before venturing into organs-on-a-chip (now also called tissue-chips) and developing a disease-on-a-chip (15), specifically for carcinomas, and more recently, a gradient-on-a-chip to reproduce microenvironmental heterogeneity (16). We have developed these systems to study the importance of the exquisite organization of the cell nucleus in sensing external conditions and controlling normal and cancerous cell behaviors. The model for the initial development of 3D cell culture methods has been the mammary gland, and it has been our main model of study throughout the years. With this model we present how, using standard 3D cell culture, we have revealed the importance of nuclear organization in controlling cell behavior and the interaction between the ECM and the cell nucleus. We further discuss how technology has been essential to produce microphysiological systems that recently allowed us to explore cell and nuclear functions within the physical constraints of an organ and by considering tissue heterogeneity.
3D culture to reproduce tissue architecture
The relationship between structure and function in tissues is one of the oldest observations since the invention of microscopy. In 3D cell culture that is physiologically relevant, the primary goal should be to reproduce tissue architecture, namely the organization of cells in a 3D structure like in vivo. For instance, a glandular epithelium is usually monolayered with luminal cells polarized thanks to tight junctions at their lateroapical membranes and hemidesmosomes at their basal membrane (against portions of a basement membrane; Figure 1A). Epithelial cells might be in contact with myoepithelial cells as it is the case in the breast; however, depending on the questions asked, it is often not necessary to coculture myoepithelial and luminal cells. An appropriate matrix support (e.g., laminin 111) and luminal cells have been sufficient so far to study the relationship between ECM and cell nucleus in the breast luminal epithelium, most likely because, in the presence of basement membrane components, breast epithelial cells in culture may acquire features of myoepithelial cells at their basal pole and features of luminal cells at their apical pole (7). With such simple system, as illustrated in this first part of progress report we have demonstrated the importance of nuclear organization in directing cell phenotype and determined apical polarity, under the control of microenvironment-nucleus interaction, as the earliest known architectural barrier against cancer onset.
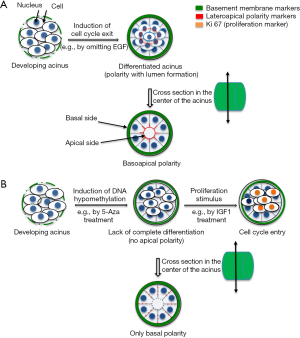
Importance of 3D cell culture for the study of the organization of the cell nucleus
The organization of the cell nucleus has been a fascinating story. It is still a main part of the pathological evaluation and diagnosis of cancers through simple light microscopy observation of hematoxylin & eosin staining that shows heterogeneity in nuclear shape and size, and in chromatin density. It is also at the center of major scientific developments thanks to approaches that permitted the discovery of noncoding RNAs and epigenetic modifications. However, only 3D cell culture could help demonstrate that the distribution of nuclear proteins mattered to maintain normal tissue differentiation. Following the initial observation that the structural protein, Nuclear Mitotic Apparatus (NuMA), displayed remarkable differences in distribution between normal and cancer phenotypes in vivo (18,19), we used basement membrane-induced breast glandular structures with epithelial polarity axis in 3D cell culture to study the influence of the distribution of NuMA on differentiation. Forcing the homogeneous distribution of the protein in the cell nucleus, both during the 10-day differentiation process and in already differentiated structures instead of the focal distribution pattern observed in normally differentiated cells, was sufficient to prevent the formation of the polarity axis (20). Whereas altering NuMA in nondifferentiated cells, like in standard 2D culture, did not show remarkable phenotypic changes. In the early days of the epigenetic revolution, we further strengthened the importance of nuclear organization to control phenotype by preventing DNA methylation during glandular differentiation, which also impaired the formation of a complete polarity axis (7) (Figure 1B).
These early findings support the idea proposed by others in the cell nucleus field that upon normal development of a tissue, the ultimate differentiation stage is locked in place at the gene transcription level by a specific nuclear organization (21). It is now known that the lock is epigenetic, but epigenetic modifications are not the only features of sustainable differentiation; a higher order nuclear organization that can only be recapitulated with 3D cell culture and includes chromatin loop formation in relation to structures such as the lamina, the nucleolus and possibly nuclear speckles seems necessary to achieve proper phenotypic control (22-24).
Extracellular matrix, cell nucleus and apical polarity relationship
One of the most difficult structural features of epithelial tissues to reproduce is a complete polarity axis. Basement membrane signaling permits the establishment of basal polarity; however, apical polarity defined by the presence of tight junctions at the lateroapical side of cell-cell contacts has been often underestimated, and the apical distribution of the Golgi apparatus has been used as a surrogate although it is not a complete representation of polarity. To display the apical pole of polarity, cells need to be properly managed already during maintenance in 2D culture as we illustrated in a previous publication (25), and the presence of collagen IV in the matrix in 3D culture appears to play an important role (8).
With the polarity-capable breast epithelial HMT-3522 S1 cell line in 3D culture, we established that the presence of apical polarity prevents cells from exiting quiescence, even when using a strong proliferation stimulant like insulin growth factor (IGF)-1. Indeed, only when cells do not display apical polarity, can they be pushed into the cell cycle either by IGF-1 or by altering the organization of the cell nucleus (17,26) (Figure 1B). These findings, along that of other teams, promoted the acceptance by the breast cancer research community that the alteration of polarity is necessary for cancer onset (27). The contribution of 3D cell culture to the demonstration of the importance of polarity in maintaining tissue homeostasis has placed differentiation models with non-neoplastic cells at the forefront of the work necessary to develop research on the primary prevention of differentiation disorders, like cancers (28).
Basement membrane-based matrices for 3D cell culture have been very useful to study the physiologically relevant differentiation of epithelia. However, such drip or embedded culture systems are mainly of interest for tissues that form spheroidal structures, like exocrine glandular tissues and certain forms of cancer. Combining forces with engineers for the microfabrication of cell culture systems with a more controlled environment has exponentially broadened the spectrum of physiologically relevant models.
The era of tissue-chips
Reproducing portions of an organ or a specific tissue using technology (e.g., in the matrix design and the microfabrication of culture platforms) has revolutionized 3D cell culture. These methods were initially developed with the goal of producing organs-on-a-chip for pharmacological purposes, hence leading to the microfluidic platforms for lung, vascular, liver and kidney models (29). Organs-on-a-chip do not necessarily mean that all tissue types in an organ are represented, but rather there should be a minimal organization and cell types necessary to ensure tissue function and test the response to therapeutic drugs. Some of the proposed models for organs-on-a-chip do not reproduce the architecture of the tissues that represent the function of the organ of interest, which we hypothesized to be responsible for the lack of sustained differentiation of these tissues (30). We anticipated that the growth of the era of cell culture on-a-chip would certainly also benefit the study of diseases. Thus, we designed the prototypes for the disease-on-a-chip (15) and gradient-on-a-chip (16) that mimic geometrical constraints as well as microenvironmental heterogeneity under which disorders like carcinomas develop. We have used these models to further the study of the influence of the microenvironment on nuclear organization for the control of tissue phenotype as illustrated in the next two sections.
Mimicry of tissue geometry
The most prevalent carcinomas involve glandular systems, like in the breast, prostate and pancreas. In these systems tumors initially grow within the confines of a ductal structure. The demonstration of the importance of mechanical constraints in the matrix to control cancer cell phenotype (31) provided new information on how sensitive cells are to physical parameters in their environment, notably ECM tension, interstitial fluid pressure and shear stress. However, there are also mechanical constraints linked to the specific shape of a tissue, to which we refer as tissue geometry. Hence, we asked whether the curved geometry of the ductal epithelium in which carcinomas develop was leading to architectural constraints that would also affect tumor behavior.
To fabricate a curved geometry, laser micromachining seemed ideal. It can be done on substrata amenable to cell culture like plastic, paper and sufficiently resilient gels. We opted for a hemichannel structure rather than a complete channel to facilitate the deposition of cells and the study on the same device of tumors cultured on a curved or on a flat geometry (Figure 2). The idea was to culture tumors in the presence of ductal structures covered by differentiated (polarized) non-neoplastic epithelial cells to reproduce cancer development conditions as in vivo. Surprisingly, the curved geometry lined by one layer of epithelial cells made a significant difference in terms of tumor sensitivity to treatments with cytotoxic anticancer drugs (15). Under these geometrical constraints we investigated possible changes in tumor morphology that might coincide with changes in drug sensitivity. Only the nucleus of the cell, not the cell itself, displayed significant alterations in morphometry (Figure 2). These results are in line with the observation by others that the expression of p53, a protein essential for the response to drug induced-cell death, is related to nuclear morphometry, such as shape (32). This striking observation of the link between tissue geometry and cancer cell phenotype raises the question of the importance of the type of 3D cell culture system to utilize in drug development and testing. Indeed, it is increasingly recognized that anticancer drug investigations in 2D cell culture are obsolete when considering results obtained with cancer cells induced to form tumors in culture. Yet, with the demonstration of the importance of not only matrix stiffness, but also geometric features of the environment in controlling cancer cell behavior, it seems that the type of 3D cell culture used to produce cancer nodules will have to be further refined. When we started working with hemichannels (33), other scientists published systems with a closed ductal structure that are likely to be also of great interest to further study certain aspects of the interaction between the microenvironment and cancer phenotypes (34).
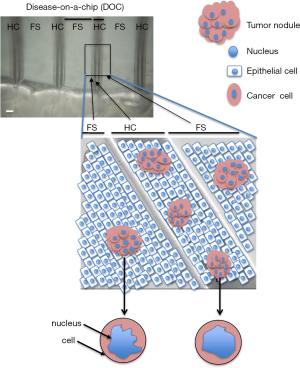
Production of controlled microenvironments
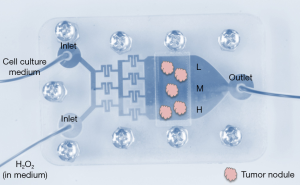
Microphysiological systems may potentially recapitulate all environmentally relevant physiological parameters of tissue homeostasis and disease development such as dynamic nutrient flow, mechanical forces in the tissues, and gradients of soluble molecules occurring between the blood and other tissues. Knowledge is steadily improving regarding the influence of physical properties of the ECM, such as stiffness and porosity, on cell behavior in development and differentiation (35,36). An external mechanical stimulus may trigger intracellular responses and alter cellular communication (37,38). Examples of external stimulus include shear stress from the vasculature (39), topographical features of the ECM such as the arrangement of nanoscale collagen fibrils into microscale collagen fibers that remodels during disease conditions (40,41), and changes in ECM stiffness (42,43). Moreover, evidence of the influence of fibroblast-ECM interaction on cancer progression (44) suggests that the interaction between distinct cell types might be essential to fully grasp changes in cellular response under varying microenvironmental conditions (45,46). Microfluidic platforms have often been chosen to better understand the interaction between stromal cells and the tissue responsible for organ functions while manipulating physical and mechanical aspects of the ECM (47,48). Yet, these platforms did not reproduce the heterogeneity that constitutes a major trait of the microenvironment in vivo. We have recently designed and tested a microfluidic system that reproduces concentration gradients to study the impact of microenvironmental mediators of oxidative stress on cancer progression (16).
Initially researchers had the idea to produce gradients for cell culture because of heterogeneity in oxygen consumption. The levels of oxygen fluctuate greatly within organs, with definite consequences for the treatment of diseases such as cancer (49-51). The kinetics of cellular oxygen consumption and dynamic changes in oxygen concentrations in the fluid surrounding the cells have been reproduced in 3D culture thanks to microfluidic systems that monitor and control oxygen delivery over time (52-54). In addition to periods of oxygenation and hypoxia, oxidative stress generated by microenvironmental reactive oxygen species (ROS) also influences normal and diseased cell phenotypes, including proliferation and differentiation (55). The heterogeneous distribution of ROS in the stroma depends on differential needs by the stromal cells such as fibroblasts and immune cells and the availability of ROS clearing antioxidant enzymes (56,57). To recreate heterogeneity in ROS in the microenvironment, we used a microfluidic system in which the gradient generated via a maze of channels results in the delivery of decreasing concentrations of H2O2 along a series of parallel microchannels (16). Thus, tumors embedded in collagen 1 would be in contact with different amounts of ROS depending on their location in the same culture chamber (Figure 3). The impact of ROS on the phenotype of preinvasive breast tumors was assessed following acute exposure. Since treatment was only four hours, we used nuclear morphometry as evidence of preliminary signs of phenotypic impact. Indeed, in collaboration with the Moghe laboratory we had shown earlier that changes in nuclear organization constitute one of the earliest markers of phenotypic switch (58). With the gradient-on-a-chip we showed that ROS modified nuclear morphometry in a way that suggests transition toward a more aggressive phenotype (with increased nuclear size and decreased circularity) (59,60). Importantly, the concentration at which ROS had an impact on nuclear morphometry depended on the stiffness of the matrix, indicating that the combination and monitoring of several microenvironmental factors involved in cancer progression is an important step to achieve with microphysiological systems.
Conclusions on the impact of tissue-chips for the understanding of microenvironment-nucleus interaction and future directions
Recreating a controlled tissue microenvironment on-a-chip has been a significant and recent revolutionary step in biological research. We have presented examples from our laboratory in which microphysiological systems have served our quest for a better understanding of the interaction between microenvironment and cell nucleus to control phenotypes. We have shown that the ECM controls nuclear organization and, in turn, cell phenotypes, initially with simple 3D culture models. But, as we illustrated above, the full extent of mechanical influence that includes tissue geometry required micromachined tissue-chips, and the full extent of chemical influence that includes microenvironmental heterogeneity required gradients created with microfluidics. We have used nuclear organization as a marker of phenotypic changes because this architectural feature is a very sensitive and early marker annunciating epigenetic modifications. Yet, sophisticated culture systems are usually lacking the possibility to easily assess changes in gene transcription. Indeed, these miniaturized systems only provide a small number of cells and mostly prevent cell removal without destruction of the tissue architecture. It appears that the gradient-on-a-chip with an open culture chamber that we recently developed and tested, and in which cell cultures are performed on a cellulose acetate substratum, might provide one of the solutions for easier assessment of gene expression (16).
Evolving 3D cell culture models have been instrumental to understand how nuclear morphology controls cell phenotypes. The impact of changes in nuclear morphometry on gene expression is thought to occur in part via geometrical modifications, with chromosomal organization as an intermediate (mediator) step (61). The influence of the nuclear lamina, itself partly directing nuclear morphometry and peripheral heterochromatin, is becoming increasingly understood in the control of phenotypes via chromatin organization (62); however, additional proteins that organize the epigenome and are located within the cell nucleus require more attention in the control of phenotype, like for instance the protein NuMA revealed by our work with 3D models of breast epithelial differentiation.
Literature reports show that the role of nuclear morphology in controlling cell phenotypes cannot be dissociated from the cell nucleus response to microenvironmental impact. Although chemical signaling to the cell nucleus is relatively well understood thanks to the identification of pathways that control the epigenome, mechanotransduction mechanisms at the cell nucleus level remain a big challenge that will require more sophisticated cell culture tools to influence the physical link within cells, including fabrication methods to control networks of matrix fibers in 3D. Indeed, both passive organization of the cytoskeleton and active stress fields might be involved in nuclear morphometric alterations and positioning depending on the cell type (63), and nuclear alterations seem to respond to the direction and the duration of mechanical stress (64). Like we have shown with the breast tumors growing within curved surfaces, the geometry of the microenvironment is also important to obtain a nuclear morphology and a phenotype of cardiomyocytes similar to in vivo, as shown with microridges engineered in the microenvironment of these cells (65).
Recent progress with biomimetic ECM has allowed for fine-tuned control of the microporosity of the matrix environment necessary for isotropic or for anisotropic growth in the cell population (66). Similarly, in order to better understand how nuclear morphometry responds to the ECM, a controlled microenvironment for experiments has been envisioned with, for instance, studies that compare substrates containing (or not) biological cues and prism micropillars of different sizes (67), hence revealing which physical parameters of the nucleus is most responsive depending on biological stimulus, length of stimulus and mechanical constraints of the stimulus. Techniques such as cell microharpooning might also help better grasp nucleoskeletal coupling in mechanotransduction (68). However, progress needs to be made to better measure forces at the cell nucleus level within a population of cells (reproduced in 3D culture), since recent work has revealed that nuclear stiffness distinguishes cancer cell lineages and cancer and noncancer cells (69). Finally, it is likely, that for a better translational power, work performed on the cell nucleus with sophisticated tissue chips for preclinical and clinical applications will benefit from emerging machine learning computational approaches (70).
A focus on mechanotransduction to further study the relationship between the microenvironment and the cell nucleus is legitimate in light of the increasing number of diseases, like progeria, cancer, muscular dystrophy, lipodystrophies, and certain types of cardiomyopathies and neurological disorders, that are becoming linked to a dysregulation of this mechanism. Although changes in nuclear morphometry are related to these diseases, mechanotransduction pathways remain to be fully deciphered. It has been suggested that pressure differences across the nuclear envelop linked to microtubules and actin filaments might be implicated in influencing nuclear morphology (71). Furthermore, mechanical stimuli direct the translocation of transcription factors to the cell nucleus (72,73), suggesting that altered (or lack of) stimuli are likely to affect the epigenetic control of cellular homeostasis. The importance of the lack of mechanical stimulus in disorders is demonstrated in cells transfected with lamin A Delta50 that reduces the response of the nuclear lamina to stress induced by flow field and thus, leads to an absence of adaptation to the flow; this phenomenon has been proposed to result in cardiovascular disease development (74). In cancer, mechanotransduction-linked epigenetic modifications are only part of the equation, and it is emphasized that alterations in gene copy number and DNA are also linked to mechanotransduction through mechanisms that are still not elucidated (75). We envision that tissue-chips with controlled microenvironment and advanced biosensing will help further elucidate the mechanisms underlying mechanosensitive diseases both at the cytoskeleton and the cell nucleus levels.
In conclusion, with raising awareness that the use of animal models for raw material and drug testing is not the sole solution anymore, and with repeated evidence that animal studies are not representative of human systems (76-78), there is an increasing demand for the development of in vitro models that mimic the physiology of human body. Nevertheless, to gain full acceptance of microphysiological systems by the scientific community several challenges need to be overcome. Kits for extraction of cellular material from small quantities of cells are becoming available, which will make working with miniature cultures easier, but issues remain regarding the need for variations in scaffolding material depending on the scientific query and the facility with which microscopy can be performed. Ultimately, for reproducibility and comparison across teams, certain protocols in 3D cell culture will have to be standardized. The importance of such standardization cannot be over-emphasized for drug testing and screening. In the meantime, as there can be almost as many devices as there are scientific queries, a plethora of microphysiological systems will be developed, and biologists will keep being amazed at the possibilities provided by engineers to refine cell culture.
Acknowledgments
We thank Manuel Ochoa and Rahim Rahimi for providing pictures of on-a-chip systems shown in Figures 2 and 3.
Funding: Support for tissue-chip work in the Lelièvre laboratory presented in this manuscript include grants from the Congressionally-Directed Medical Research Program/Breast Cancer Research Program (W81XWH-09-1-0354 and W81XWH-17-1-0250 to SAL).
Footnote
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/mps.2018.11.02). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Schwarz RI, Bissell MJ. Dependence of the differentiated state on cellular environment: modulation of collagen synthesis on tendon cells. Proc Natl Acad Sci U S A 1977;74:4453-7. [Crossref] [PubMed]
- Enami J, Nandi S. Secretion of casein in culture of mouse mammary epithelial cells on floating collagen gels. J Dairy Sci 1978;61:729-32. [Crossref] [PubMed]
- Emerman JT, Burwen SJ, Pitelka DR. Substrate properties influencing ultrastructural differentiation of mammary epithelial cells in culture. Tissue Cell 1979;11:109-19. [Crossref] [PubMed]
- Barcellos-Hoff MH, Aggeler J, Ram TG, et al. Functional differentiation and alveolar morphogenesis of primary mammary cultures on reconstituted basement membrane. Development 1989;105:223-35. [PubMed]
- Petersen OW, Rønnov-Jessen L, Howlett AR, et al. Interaction with basement membrane served to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci U S A 1992;89:9064-8. [Crossref] [PubMed]
- Gudjonsson T, Rønnov-Jessen L, Villadsen R, et al. Normal and tumor-derived myoepithelial cells differ in their ability to interact with luminal breast epithelial cells for polarity and basement deposition. J Cell Sci 2002;115:39-50. [PubMed]
- Plachot C, Lelièvre SA. DNA methylation control of tissue polarity and cellular differentiation in mammary epithelium. Exp Cell Res 2004;298:122-32. [Crossref] [PubMed]
- Plachot C, Chaboub LS, Adissu HA, et al. Factors necessary to produce basoapical polarity in human glandular epithelium formed in conventional and high-throughput three-dimensional culture: example of the breast epithelium. BMC Biol 2009;7:77. [Crossref] [PubMed]
- Bissell MJ, Hall HG, Parry G. How does the extracellular matrix direct gene expression? J Theor Biol 1982;99:31-68. [Crossref] [PubMed]
- Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol 2009;10:75-82. [Crossref] [PubMed]
- Athirasala A, Hirsch N, Buxboim A. Nuclear mechanotransduction: sensing the force from within. Curr Opin Cell Biol 2017;46:119-27. [Crossref] [PubMed]
- Jorgens DM, Inman JL, Wojcik M, et al. Deep nuclear invaginations are linked to cytoskeletal filaments – integrated bioimaging of epithelial cells in 3D culture. J Cell Sci 2017;130:177-89. [Crossref] [PubMed]
- Lelièvre SA, Bissell MJ. Communication between the cell membrane and the nucleus: role of protein compartmentalization. J Cell Biochem Suppl 1998;30-31:250-63. [Crossref] [PubMed]
- Fogg PCM, O’Neill JS, Dobrzycki T, et al. Class IIa Histone Deacetylases Are Conserved Regulators of Circadian Function. J Biol Chem 2014;289:34341-8. [Crossref] [PubMed]
- Vidi PA, Maleki T, Ochoa M, et al. Disease-on-a-chip: mimicry of tumor growth in mammary ducts. Lab Chip 2014;14:172-7. [Crossref] [PubMed]
- Chittiboyina S, Rahimi R, Atrian F, et al. Gradient-on-a-chip with reactive oxygen species reveals thresholds in the nucleus response of cancer cells depending on the matrix environment. ACS Biomater Sci Eng 2018;4:432-45. [Crossref]
- Yue S, Cárdenas-Mora JM, Chaboub LS, et al. Label-free analysis of breast tissue polarity by Raman imaging of lipid phase. Biophys J 2012;102:1215-23. [Crossref] [PubMed]
- Lelièvre SA, Weaver VM, Nickerson JA, et al. Tissue phenotype depends on reciprocal interactions between the extracellular matrix and the structural organization of the nucleus. Proc Natl Acad Sci U S A 1998;95:14711-6. [Crossref] [PubMed]
- Knowles DW, Sudar D, Bator-Kelly C, et al. Automated local bright feature image analysis of nuclear protein distribution identifies changes in tissue phenotype. Proc Natl Acad Sci U S A 2006;103:4445-50. [Crossref] [PubMed]
- Abad PC, Lewis J, Mian IS, et al. NuMA Influences Higher Order Chromatin Organization in Human Mammary Epithelium. Mol Biol Cell 2007;18:348-61. [Crossref] [PubMed]
- Smith CM, Haimberger ZW, Johnson CO, et al. Heritable chromatin structure: mapping memory in histones H3 and H4. Proc Natl Acad Sci U S A 2002;99:16454-61. [Crossref] [PubMed]
- Jing H, Vakoc CR, Ying L, et al. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell 2008;29:232-42. [Crossref] [PubMed]
- Lelièvre SA. Contributions of extracellular matrix signaling and tissue architecture to nuclear mechanisms and spatial organization of gene expression control. Biochim Biophys Acta 2009;1790:925-35. [Crossref] [PubMed]
- Battistelli C, Busanello A, Maione R. Functional imterplay between MyoD and CTCF in regulating long-range chromatin interactions during differentiation. J Cell Sci 2014;127:3757-67. [Crossref] [PubMed]
- Vidi PA, Bissell MJ, Lelièvre SA. Three-Dimensional culture of human breast epithelial cells: the how and the why. Methods Mol Biol 2013;945:193-219. [Crossref] [PubMed]
- Chandramouly G, Abad PC, Knowles DW, et al. The control of tissue architecture over nuclear organization is crucial for epithelial cell fate. J Cell Sci 2007;120:1596-606. [Crossref] [PubMed]
- Eccles SA, Aboagye EO, Ali S, et al. Critical research gaps and translational priorities for the successful prevention and treatment of breast cancer. Breast Cancer Res 2013;15:R92. [Crossref] [PubMed]
- Vidi PA, Leary J, Lelièvre SA. Building risk-on-a-chip models to improve breast cancer risk assessment and prevention. Integr Biol (Camb) 2013;5:1110-8. [Crossref] [PubMed]
- Selimović S, Dokmeci MR, Khademhosseini A. Organs-on-a-chip for drug discovery. Curr Opin Phamracol 2013;13:829-33. [Crossref] [PubMed]
- Lelièvre SA, Kwok T, Chittiboyina S. Architecture in 3D cell culture: an essential feature for in vitro toxicology. Toxicol In Vitro 2017;45:287-95. [Crossref] [PubMed]
- Northcott JM, Dean IS, Mouw JK, et al. Feeling Stress: the mechanics of cancer progression and aggression. Front Cell Dev Biol 2018;6:17. [Crossref] [PubMed]
- Mijovic Z, Kostov M, Mihailovic D. Correlation of nuclear morphometry of primary melanoma of the skin with clinicopathological parameters and expression of tumor suppressor proteins (p53and p16INK4a) and bcl-2 oncoprotein. J BUON 2013;18:471-6. [PubMed]
- Grafton MMG, Wang L, Vidi PA, et al. Breast on-a-chip: mimicry of the channeling system of the breast for development of theranostics. Integr Biol (Camb) 2011;3:451-9. [Crossref] [PubMed]
- Sung KE, Yang N, Pehlke C, et al. Transition to invasion in breast cancer: a microfluidic in vitro model enables examination of spatial and temporal effects. Integr Biol (Camb) 2011;3:439-50. [Crossref] [PubMed]
- O’Brien FJ, Harley BA, Yannav IV, et al. The effect of pore size on cell adhesion in collagen GAG-scaffolds. Biomaterials 2005;26:433-41. [Crossref] [PubMed]
- Peyton SR, Putnam AJ. Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J Cell Physiol 2005;204:198-209. [Crossref] [PubMed]
- Li YS, Haga JH, Chien S. Molecular basis of the effects of the shear stress on vascular endothelial cells. J Biomech 2005;38:1949-71. [Crossref] [PubMed]
- van der Meer AD, Poot AA, Feijen J, et al. Analyzing shear stress-induced alignment of actin filaments in endothelial cells with a microfluidic assay. Biomicrofluidics 2010;4:11103. [Crossref] [PubMed]
- Price GM, Wong KHK, Truslow JG, et al. Effect of mechanical factors on the function of engineered human blood microvessels in microfluidic collagen gels. Biomaterials 2010;31:6182-9. [Crossref] [PubMed]
- Birk DE, Nurminskaya MV, Zycband EI. Collagen fibrillogenesis in situ: fibril segments undergo post-depositional modifications resulting in linear and lateral growth during matrix development. Dev Dyn 1995;202:229-43. [Crossref] [PubMed]
- Canty EG, Lu Y, Meadows RS, et al. Coalignment of plasma membrane channels and protrusions (fibripositors) specifies the parallelism of tendon. J Cell Biol 2004;165:553-63. [Crossref] [PubMed]
- Wells RG. The role of matrix stiffness in regulating cell behavior. Hepatology 2008;47:1394-400. [Crossref] [PubMed]
- Baker EL, Bonnecaze RT, Zaman MH. Extracellular matrix stiffness and architecture govern intracellular rheology in cancer. Biophys J 2009;97:1013-21. [Crossref] [PubMed]
- Nishioka M, Venkatesan N, Dessale K, et al. Fibrobalst-epithelial cell interactions drive epithelil-mesenchymal transition differently in cells from normal and COPD patients. Respir Res 2015;16:72. [Crossref] [PubMed]
- Eckes B, Zweers MC, Zhang ZG, et al. Mechanical tension and integrin alpha 2 beta 1 regulate fibroblast functions. J Investig Dermatol Symp Proc 2006;11:66-72. [Crossref] [PubMed]
- Menon NV, Chuah YJ, Cao B, et al. A microfluidic co-culture system to monitor tumor-stromal interactions on a chip. Biomicrofluidics 2014;8:064118 [Crossref] [PubMed]
- Zhang Y, Park S, Liu K, et al. A surface topography assisted droplet manipulation platform for biomarker detection and pathogen identification. Lab Chip 2011;11:398-406. [Crossref] [PubMed]
- Sundararaghavan HG, Monteiro GA, Firestein BL, et al. Neurite growth in 3D collagen gels with gradients of mechanical properties. Biotechnol Bioeng 2009;102:632-43. [Crossref] [PubMed]
- Sutherland RM, Ausserer WA, Murphy BJ, et al. Tumor hypoxia and heterogeneity: challenges and opportunities for the future. Semin Radiat Oncol 1996;6:59-70. [Crossref] [PubMed]
- Pries AR, Secomb TW. Origins of heterogeneity in tissue perfusion and metabolism. Cardiovasc Res 2009;81:328-35. [Crossref] [PubMed]
- Rundqvist H, Johnson RS. Tumor oxygenation: implications for breast cancer prognosis. J Intern Med 2013;274:105-12. [Crossref] [PubMed]
- Uchida H, Sato A, Miyayama A, et al. Generation of an oxygen gradient in a microfluidic device and cellular analysis in hypoxia. Adv Biomed Eng 2013;2:143-9. [Crossref]
- Lo JF, Wang Y, Eddington DT, et al. Quantitative and temporal control of oxygen microenvironment at the single islet level. J Vis Exp 2013;81:e50616 [PubMed]
- Sové RJ, Fraser GM, Goldman D, et al. Finite element model of oxygen transport for the design of geometrically complex microfluidic devices used in biological studies. PLoS One 2016;11:e0166289 [Crossref] [PubMed]
- Chittiboyina S, Bai Y, Lelièvre SA. Microenvironment-cell nucleus relationship in the context of oxidative stress. Front Cell Dev Biol 2018;6:23. [Crossref] [PubMed]
- Kim IS, Zhang XHF. One microenvironment does not fit all: heterogeneity beyond cancer cells. Cancer Metastasis Rev 2016;35:601-29. [Crossref] [PubMed]
- de Sá PL Junior, Câmara DAD, Porcacchia AS, et al. The roles of ROS in cancer heterogeneity and therapy. Oxid Med Cell Longev 2017;2017:2467940 [PubMed]
- Vega SL, Liu E, Arvind V, et al. High-content image informatics of the structural nuclear protein NuMA parses trajectories for stem/progenitor cell lineages and oncogenic transformation. Exp Cell Res 2017;351:11-23. [Crossref] [PubMed]
- Veltri RW, Christudass CS, Isharwal S. Nuclear morphometry, nucleomics and prostate cancer progression. Asian J Androl 2012;14:375-84. [Crossref] [PubMed]
- Vedam VK, Boaz K, Natarajan S. Prognostic efficacy of nuclear morphometry at invasive front of oral squamous cell carcinoma: an image analysis microscopic study. Anal Cell Pathol (Amst) 2014;2014:247853 [Crossref] [PubMed]
- Uhler C, Shivashankar GV. Geometric control and modeling of genome reprogramming. Bioarchitecture 2016;6:76-84. [Crossref] [PubMed]
- Dobrzynska A, Gonzalo S, Shanahan C, et al. The nuclear lamina in health and disease. Nucleus 2016;7:233-48. [Crossref] [PubMed]
- Bray MA, Adams WJ, Geisse NA, et al. Nuclear morphology and deformation in engineered cardiac myocytes and tissues. Biomaterials 2010;31:5143-50. [Crossref] [PubMed]
- Nathan AS, Baker BM, Nerurkar NL, et al. Mechano-topographic modulation of stem cell nuclear shape on nanofibrous scaffolds. Acta Biomater 2011;7:57-66. [Crossref] [PubMed]
- Jain A, Hasan J, Desingu PA, et al. Engineering an in vitro organotypic model for studying cardiac hypertrophy. Colloids Surf B Biointerfaces 2018;165:355-62. [Crossref] [PubMed]
- Canadas RF, Ren T, Tocchio A, et al. Tunable anisotropic networks for 3-D oriented neural tissue models. Biomaterials 2018;181:402-14. [Crossref] [PubMed]
- Antmen E, Ermis M, Demirci U, et al. Engineered natural and synthetic polymer surfaces induce nuclear deformation in osteosarcoma cells. J Biomed Mater Res B Appl Biomater 2018; [Epub ahead of print]. [Crossref] [PubMed]
- Fedorchak G, Lammerding J. Cell microharpooning to study nucleo-cytoskeletal coupling. Methods Mol Biol 2016;1411:241-54. [Crossref] [PubMed]
- Ermis M, Akkaynak D, Chen P, et al. A high throughput approach for analysis of cell nuclear deformability at single cell level. Sci Rep 2016;6:36917. [Crossref] [PubMed]
- Carleton NM, Lee G, Madabhushi A, et al. Advances in the computational and molecular understanding of the prostate cancer cell nucleus. J Cell Biochem 2018;119:7127-42. [Crossref] [PubMed]
- Kim DH, Li B, Si F, et al. Volume regulation and shape bifurcation in the cell nucleus. J Cell Sci 2015;128:3375-85. [Crossref] [PubMed]
- Cardoso AC, Pereira AHM, Ambrosio ALB, et al. FAK forms a complex with MEF2 to couple biomechanical signaling to transcription in cardiomyocytes. Structure 2016;24:1301-10. [Crossref] [PubMed]
- Monticelli M, Jokhun DS, Petti D, et al. Localized mechanical stimulation of single cells with engineered spatio-temporal profile. Lab Chip 2018;18:2955-65. [Crossref] [PubMed]
- Philip JT, Dahl KN. Nuclear mechanotransduction: response of the lamina to extracellular stress with implications in aging. J Biomech 2008;41:3164-70. [Crossref] [PubMed]
- Discher DE, Smith L, Cho S, et al. Matrix mechanosensing: From scaling concepts in ‘Omics data to mechanisms in the nucleus, regeneration, and cancer. Annu Rev Biophys 2017;46:295-315. [Crossref] [PubMed]
- Hackam DG, Redelmeier DA. Translation of research evidence from animals to humans. JAMA 2006;296:1731-2. [Crossref] [PubMed]
- Sena ES, van der Worp HB, Bath PMW, et al. Publication Bias in Reports of Animal Stroke Studies Leads to Major Overstatement of Efficacy. PLoS Biology 2010;8:e1000344 [Crossref] [PubMed]
- Hartung T. Food for thought look back in anger – what clinical studies tell us about preclinical work. ALTEX 2013;30:275-91. [Crossref] [PubMed]
Cite this article as: Lelièvre SA, Chittiboyina S. Microphysiological systems to study microenvironment-cell nucleus interaction: importance of tissue geometry and heterogeneity. Microphysiol Syst 2018;2:12.


